

Lipids are insoluble in water, so they are synthesized in a membrane bilayer that makes a separate phase, like a layer of oil forming on water (Figure 1). Since most lipid-synthesizing enzymes are localised to just one membrane, the endoplasmic reticulum in eukaryotic cells or the inner membrane of bacteria, there is a major question: how can lipids transfer to another membrane? A method that might work on the face of it is traffic of vesicles made
from membrane patches. However, the data do not support this. Traffic of vesicles in eukaryotic cells cannot provide lipids to organelles such as lipid droplets, mitochondria and peroxisomes. Even more definitively, the vast majority of cells that require lipid traffic are bacteria with 2 membranes (such as E. coli), which have no membrane vesicles. This means that life on earth solved the question of lipid traffic long before vesicular traffic evolved. This paper is part of a trend to return the field of lipid transfer back to it’s roots.

Lipids are trafficked to their intended location by Lipid Transfer Proteins (LTPs). These come in two types: ‘boxy’ LTPs that resemble lidded boxes that can select cargo lipids for transport across water filled gaps such as the cytosol, like an oil barrel that can be filled with crude oil; and ‘groovy’ LTPs that resemble half-pipes through which large quantities of cargo can flow (Figure 2). The pipes are relative new discoveries that suggest the intriguing possibility that lipids can simply slide from one membrane to another at so-called “membrane contact sites” where membranes come closest together. The typical gaps at these sites is 15-20 nm, which matches the lengths of the known half-pipes.
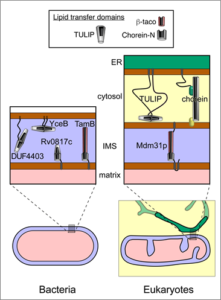
The featured paper by Tim Levine (UCL, United Kingdom) predicts the existence of multiple bacterial homologues of the two LTPs folds known to form “groovy” LTPs inside eukaryotic cells. One fold is the tubular lipid binding (TULIP) superfamily, which have quite short hydrophobic grooves (up to 7 nm long) – too short to stretch all the way between two membranes where they contact one another. The other “groovy” fold is the Chorein-N domain found at the N-terminus of various proteins including the human protein chorein. The first description of this protein as a half-pipe in 2018 revolutionised thinking about lipid transfer. Chorein has the internal capacity to hold many lipids (>20), forms rods 20 nm long, and is localised to sites where two membranes come very close together. The featured paper shows that domains that are strongly homologous either to TULIPs or Chorein-N (and in some cases both) are widespread in bacteria (Figure 3). In addition, there are previously unknown homologues in mitochondria, presumably inherited from the bacterial ancestor of these symbiotic organelles.
This prediction of likely bacterial homologues of these LTPs points to what experiments should be done next to investigate modes of “groovy” lipid transfer in bacteria, the most simple and tractable cellular system. This can bring us a lot closer to working out how lipids reach their destinations in all cells.
Comments