

The ability to genetically mutate organisms has long been fantasized in the world of science fiction, perhaps the most famous example being the X-men franchise, where a genetic mutation can grant superhuman powers. While scientists have not yet unlocked the genetic code necessary for telekinesis, targeted genetic editing has existed since 1974 to probe and explore genetic functions. Manipulating protein levels and activities is a principal tool in understanding the functions and relationships of the molecular components in cells, but true genetic manipulation to produce a stable transgenic cell line or a true-breeding organism requires far more advanced molecular manipulation, and older techniques are hinder-some to the modern scientist in that they are time and labor intensive, requiring a scientist to screen though generations of organisms, as well as having advanced knowledge of molecular techniques.
The newest tool in the geneticist’s tool box makes use of the clustered, regularly interspaced short palindromic repeats (CRISPR)/Cas system, an endogenous adaptive immune system commonly used by bacteria and archaea to counteract “infection” by foreign DNA. This new method dramatically changes the field of molecular biology, and will allow scientists move forward with genetic experimentation in leaps and bounds.
The type II CRISPR/Cas system from Streptococcus pyogenes appears to be particularly well suited for scientists performing genomic editing experiments for a simple reason: a single protein, Cas9, is required for crRNA binding and cleavage, and the RNA components have been engineered into a single guide RNA (sgRNA). Because of this, targeting of nearly any genomic sequence is possible with these two reagents in the cells of interest, and the creation of a knockout line depends solely on DNA cleavage by the Cas endonuclease followed by non-homologous end joining (NHEJ) repair of the site by the cell.
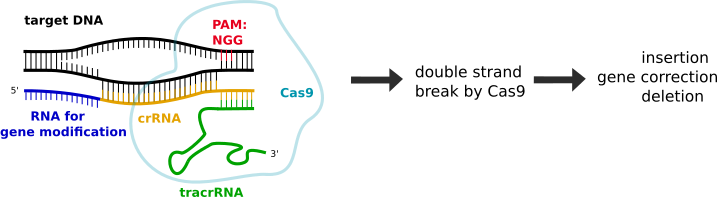
When the CRISPR system completes the transfection process, a small, random deletion is often introduced at the repair site, which likely leads to a non-functional protein sequence that can be targeted for nonsense-mediated mRNA decay. The Cas cleavage can also stimulate homology-driven repair based on a supplied DNA template, and allow for larger deletions or insertion of tags and markers, whereas older methods were unable or limited in doing so.
In their article published in BMC Molecular Biology, Estep and colleagues describe an improved method for clonal screening without selection: using immunoblot analysis, the previous method of isolating clones by performing PCR to amplify DNA followed by sequencing or restriction digestion becomes obsolete. By screening directly for protein production, genetic knockout can be confirmed, and non-specific positive signals (false positives) unrelated to protein function can be avoided. The researchers successfully used this method to rapidly generate genomic mutants.
The regulation of nonsense-mediated mRNA decay
Scientists have made quite a few breakthroughs with regards to understanding the regulatory processes of mRNA this year. In an article published in BMC Molecular Biology, Jin and colleges at Huazhong Agricultural University have advanced our understanding of the mechanisms employed for nonsense-mediated mRNA decay, the protein surveillance method that targets and breaks down non-functional mRNA
Nonsense-mediated mRNA decay clears cells of nonfunctional proteins by targeting mRNAs which contain a premature termination codon, which would otherwise be translated into a truncated protein sequence. The cellular mechanism involved in this system is not yet completely understood, though it is known that the transcription regulating protein UPF1 activates mRNA decay by recognizing abnormal translation, by using a protein complex which includes the SMG5 protein. When SMG5 is dephosphorylated it down-regulates nonsense-mediated mRNA decay.
Jin and colleagues discovered that a microRNA, miR-433, can repress the expression of SMG5, and observed a suppression in nonsense-mediated mRNA decay, presenting the possibility of new clinical treatments for various diseases which involve protein coding mutations.
Noncoding microRNAs might just be coding for something after all
The role of microRNA (very short non-coding sequences of RNA also known as miRNA) interactions within the cell is incompletely understood, and deciphering their role oftentimes requires a complex series of experiments to confirm their independent impact on cellular processes. While it is known that miRNA modulates approximately 60% of protein translation and degradation in humans, how this is accomplished is still very much a mystery.
One area of research that featured strongly in BMC Molecular Biology in 2016 was the investigation of miRNA function in organ and tumor development.
In a series of experiments on mice, Bartram and co-workers at the University of Cologne showed that miRNAs that regulates Pax8 (a transcription factor involved in regulating the genes within the stem cells that ultimately develop into epithelial organs) are also involved in hypothyroidism and end-stage renal failure.
Li and colleagues from Chongqing Medical University performed a series of experiments on human cervical tissue from cervical cancer patients and healthy controls and observed that miRNA-150 regulates the expression of FOXO4, a gene which regulates mitosis during the cell cycle and is found to increase with the progression of cervical cancer. This new information present a new possible target for cancer treatment therapies.
Both of these studies highlight the important role miRNA plays in regulating multiple aspects of the cell cycle, and emphasize how incomplete our knowledge is of these highly important but often overlooked regulatory molecules. Increasingly, however, state of the art techniques are being used to catalogue miRNA abundance in different cell and tissue types. For example, in an article published in BMC Molecular Biology earlier this year, Tonge and Gant at Keele University reported the global expression levels of all circulating miRNAs within the plasma of normal human subjects, greatly accelerating future analysis of miRNA function in plasma. By learning of the baseline expressions of human miRNAs, the researchers hope to use this library of over 500 miRNA signatures as baselines for biomarkers. miRNA biomarkers will be particularly useful for identifying organ or system specific protein deregulation or disease since miRNAs are highly specific in location and purpose.
These new discoveries in molecular biology are transforming modern genetics by allowing future genetic modeling and experimentation to be quickly and easily accomplished. Future studies will undoubtedly benefit from these new techniques and discoveries and build from them, allowing scientists to further broaden their horizons in understanding the finer details of our own species.
Comments