Within only a few years, a technology referred to as “CRISPR” has not only changed the way scientists make specific changes in DNA, but also the way people can think about treating diseases. Meanwhile, CRISPR-Cas systems in the microbial kingdom have hastened the emergence of new genome engineering toolboxes.
A new study in Genome Biology, completed by a team led by Dr. Wei Li at Institute of Zoology, Chinese Academy of Sciences, provides us with more viable Cas12a/Cpf1 orthologs, which expands our choices of CRISPR-Cas-based genome engineering, and enhances the targeting efficiencies of Cas12a enzymes via engineering and optimizing the CRISPR RNA (crRNA) scaffold.
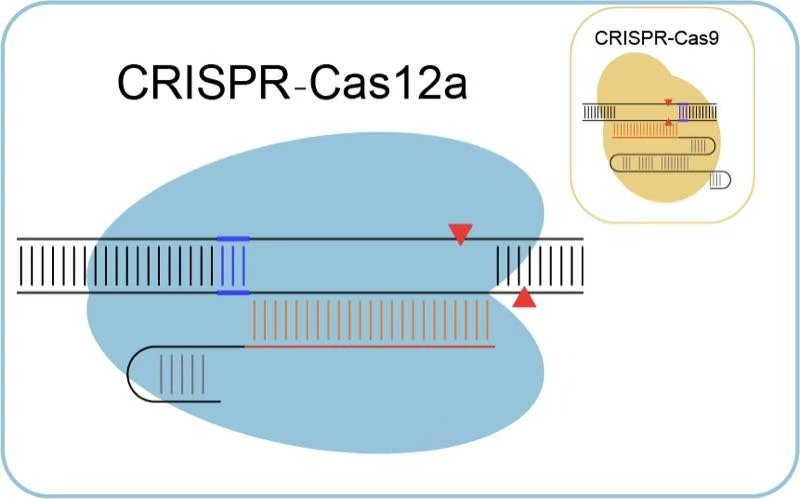
The emerging CRISPR-Cas-mediated genome editing technology is a promising area with bright prospects in fundamental researches and biomedical applications. Up to now, 6 types and over 20 subtypes of CRISPR-Cas systems have been nominated. Among them, CRISPR-Cas12a is the second type (type V) of CRISPR-Cas systems harnessed to edit mammalian genomes.
My colleagues and I “dig” the databases and identify several new CRISPR-Cas12a loci, which have been harnessed to execute mammalian genome editing functionality.
Cas12a possesses unique features, making it a mutual complementation of CRISPR-Cas9 systems. However, some disadvantages still exist and require improvement such as lower genomic coverage and less targeting efficiency.
Expanded repertoire of Cas12a toolbox
In our recent study, published in Genome Biology, my colleagues and I “dig” the databases and identify several new CRISPR-Cas12a loci, which have been harnessed to execute mammalian genome editing functionality. We demonstrate that six new Cas12a orthologs are able to edit genomes in mammalian cells.
We find that type V-A Cas12a systems are conserved, particularly in the mature crRNA sequences and second structures. Based on the conservation, we infer the conservation of their requisite PAM sequences. As our in vitro data indicated, these newly identified Cas12a nucleases utilize 5’T-rich PAM. We therefore have provided more choices for Cas12a nucleases that are able to robustly edit mammalian genomes.
Increased genomic coverage using non-canonical PAMs
We also find that some of these six Cas12a orthologs can recognize a simpler 5’-TTN PAM, which indicates a fourfold increase in genomic coverage in principle, compared with the widely used AsCas12a and LbCas12a. Moreover, in our in vitro cleavage assays, we find that, HkCas12a can utilize non-canonical PAMs (5’-YYN and 5’-TBBN-). Though less PAM sequences were determined in cell lines, we can also implement HkCas12a for mammalian genome editing using 5’-YTN and 5’-TYYN PAMs. Compared with the canonical 5’-TTN PAM, the PAMs recognized by HkCas12a increases the targeting range of HkCas12a by threefold in mammalian genomes.
Elevated targeting efficiencies of Cas12a proteins with optimized crRNA scaffolds
In our study, we observe that different crRNA scaffolds can affect the targeting efficiencies of Cas12a proteins, though in most cases they decrease or even eliminate their activities. But in rare cases, some changes in the loop region of crRNA scaffold are capable of increasing the activities of Cas12a nucleases. Successfully, we screen out one variant (crRNA4n96), which can markedly increase the Cas12a-medaited genome editing efficiencies.
Overall, we develop six new Cas12a genome editing toolboxes with increased genomic coverage, which provides us more choices in genome editing. Importantly, by engineering and optimizing the scaffold of crRNA, we enable elevated targeting efficiencies of Cas12a orthologs.
Wei Li & Fei Teng
Fei Teng is a Ph.D. candidate in Institute of Zoology, Chinese Academy of Sciences. His current research pursues the application of CRISPR-Cas systems in disease modeling and development of new genome editing toolbox.
Comments