The beauty of the CRISPR/Cas editing system is that while the single guide RNA (sgRNA), straightforward to design and synthesize, is the part that dictates the targeting properties of the whole tool, Cas9 – a stable, unchanging element – is responsible for only one thing: cutting the DNA strand in the place to which it was guided.
And we are frequently so focused on the benefits and limitations of single guide RNAs that we forget that Cas9 itself has one important restriction: it only cuts the DNA sequence recognized by sgRNA if there is a so-called protospacer adjacent motif (PAM) present next to it. PAM is a specific type of short sequence: the most commonly used wild-type Streptococcus pyogenes Cas9 (SpCas) PAM uses NGG (where N is any base). This presents an obvious obstacle for precision gene editing – sgRNAs are of limited length, and so if there is no PAM sequence close to the location in the genome that we want to modify, it may simply not be possible.
Unsurprisingly then a lot of research has been focused on extending the repertoire of PAMs recognised by Cas9 proteins.
Many faces of Cas9
As it turns out, PAM is not a conserved feature – it tends to differ for Cas9 proteins from different bacterial species. That another Cas9 can be used for genome editing has been demonstrated a few times already. In one of the early CRISPR genome editing articles in 2013, Feng Zhang, Luciano Marraffini and colleagues described a small Cas9 protein from Streptococcus thermophilus (St1Cas9). This protein is smaller than SpCas9 (3.3kb vs 4.5 kb), but has a rather complicated PAM sequence, NNAGAAW, which is even more restrictive than that of SpCas9.
Earlier this year, however, Feng Zhang and colleagues published an article in Nature describing in vivo editing performed using yet another Cas9 enzyme derived from Staphylococcus aureus (SaCas9).
SaCas9 has some interesting properties: it is as efficient an editor as SpCas9; its PAM, although longer than SpCas9’s, is quite versatile (NNGRRT); the most interesting feature of the enzyme, though, is its size – it’s only about three quarters of SpCas9’s. Why does it matter? For many applications adeno-associated viral (AAV) vectors are used for in vivo delivery. However, packaging a large SpCas9 and a single guide RNA into one vector is tricky, because the vector is barely larger (4.5kb) than the size of these constructs (4.2 kb). This is much less of an issue with smaller SaCas9.
In an article published in Genome Biology, David Bumcrot and colleagues characterize SaCas9 thoroughly in terms of its permissive PAMs. Importantly though, they also describe a design of AAV vector with SaCas9 and not one, but two single guide RNAs included. The authors speculate that with the use of different promoters (human tRNA promoters which are only 70 bp, instead of the currently used 250 bp U6 promoter) it may be possible to include even more sgRNAs within the same construct.
The search for a different Cas9 is not over yet though. In another article published in Genome Biology’s special issue on genome editing, Virginijus Siksnys and colleagues describe a method for characterization of PAMs and guide RNA requirements in newly discovered Cas9 proteins. With this method they confirm canonical PAMs of SpCas9, St1Cas9 and St3Cas9, but also identify the PAM for a novel Cas9 enzyme from Brevibacillus laterosporus (BlatCas9) and demonstrate its application for in planta editing. With this easy approach to PAM finding, any cas9-carrying bacterium is suddenly a potential source of a new biotechnological tool.
Frankenstein’s Cas9
If you’re more bioengineering-savvy and do not fancy a novel Cas9 hunt, there is another way of expanding the repertoire of PAMs – and thus the repertoire of loci in the genome that can be targeted with the CRISPR/Cas system. That is: to engineer variants of a particular Cas9 with altered PAM specificities.
This is the approach that Keith Joung’s group has taken. Last June the group published an article in Nature describing engineering of SpCas9 protein: using structural information on this enzyme, bacterial selection-based directed evolution and combinatorial design the researchers obtained a number of SpCas9 variants with different PAM requirements. They then tested two of those in zebrafish embryos and in human cells.
This work just scratches the surface of the range of PAMs that can be targeted by Cas9
Keith Joung, Harvard
Earlier this month the authors followed up with a similar study performed on SaCas9, which was published in Nature Biotechnology. You may remember that this enzyme has a fairly versatile PAM of NNGRRT. Joung and colleagues used their technique to engineer a SaCas9 variant with a NNNRRT protoadjacent motif sequence. This increased the targeting range of this nuclease two- to fourfold!
Interestingly, in their second article the researchers did not use structural information any more: the results were obtained purely by molecular evolution to modify SaCas9’s PAM. This widens the applicability of this approach, as for many novel Cas9 proteins structural information may not be available (as a matter of fact, only a few Cas9 proteins have had their structure resolved crystalographically so far).
A slightly different bioengineering approach was proposed in an article in Nature Methods in October: Scot Wolfe and colleagues described a fusion of Cas9 with a programmable DNA-binding-domain. This domain can be designed in concert with sgRNAs – and the recognition specificity is increased substantially. More importantly though, the SpCas9 fusion protein was equally efficient for a range of PAMs: NAG, NGA, NGC, and (SpCas’s canonical) NGG. One could imagine that in the future, if a nuclease can be designed in which DNA affinity defined by its recognition sequence and the fusion domain is very high, the PAM recognition site may not matter anymore at all.
The one that got away
While the main focus stays on the Cas9 nuclease – because as it is, it is the best characterized, commercially most available CRISPR-system enzyme that can be used for genome editing experiments – the attention of researchers in the field has recently diverted to another protein.
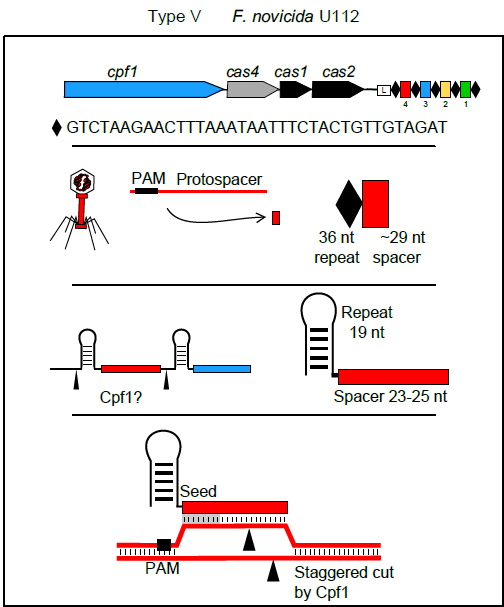
In September Feng Zhang and colleagues reported in Cell on a new single RNA-guided endonuclease of a class 2 type V system: Cpf1. A major advantage of this enzyme over Cas9 is the fact that its interaction with the CRISPR array is independent of tractRNA (Peter Fineran and colleagues provide a more detailed explanation of these differences in Genome Biology’s Research Highlight). Additionally, Cpf1’s crRNAs are much shorter than those of Cas9. All of that means that the synthesis of guide RNAs for bioengineering purposes should be much simpler, faster and cheaper than for Cas9 experiments.
Cpf1 has its own limitations though: one of them is, of course, the PAM. Its PAM requirement is TTN and CTA – interestingly, positioned on the other side of the spacer in comparison with Cas9. However, given the plethora of solutions already developed to address PAM restriction issue in Cas9, it is not a huge stretch of imagination to foresee similar approaches being used for Cpf1 in the near future.
Comments