As the human genome sequence was completed, so the deep analysis of it began in earnest with the ENCyclopedia Of DNA Elements (ENCODE) project – intended to identify all functional elements in the human genome.
The project involved a worldwide consortium of research groups and the data emerging can be accessed through public databases. BioMed Central has published a selection of the early findings.
ENCODE initiatives lead to modENCODE a project aspiring to identify the functional elements in the genomes of the model organisms Caenorhabditis elegans (nematode) and Drosophila melanogaster (fruit fly). The extension of the ‘ENCODE approach’ to other model organisms allows further biological validation of the findings coming from the human genome project.
And building on ENCODE doesn’t stop there…
Although some of the particular findings of the ENCODE project have been criticized, ENCODE-like annotations are becoming more accepted as way to advance research in model organisms.
As a result, Denis Tagu, John Colbourne and Nicolas Nègre, in their correspondence article published today in BMC Genomics, argue that the next step in the process is to understand how genetic systems react in the natural context of living ecosystems.
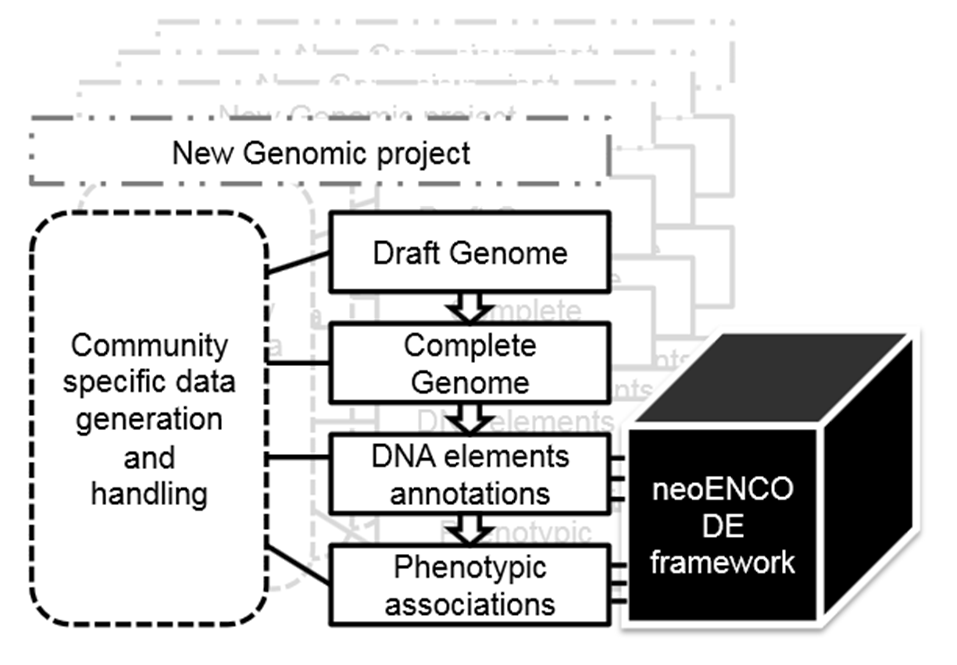
They refer to this approach as ‘neoENCODE’, the functional annotation of genomes applied to emerging and non-model organisms which are typically not amenable to large-scale forward genetics.
Unlike the organizational structure of ENCODE and modENCODE, however, Tagu and colleagues propose that neoENCODE should have an integrative approach piecing together the requirements of the communities involved.
This is not a proposal for a new consortium led by a coordinating group but for several ‘bottom-up’ initiatives within individual communities adopting, organizing and adapting their research independently. The ‘common thread’ is a shared road-map and exchanges between the various neoENCODE communities to enhance individual projects.
Openness
In submitting to BMC Genomics, the authors were keen to initiate a discussion within the community about their proposals. To facilitate this they requested that the peer review process be fully open, so that readers have access to the reviewers’ original comments.
Therefore, in an unprecedented move for BMC Genomics, (which usually operates closed peer review) the journal has published the comments that were received on an earlier version of the manuscript at the end of the correspondence piece.
In so doing, the peer reviewers themselves have started the conversation about neoENCODE and readers can hear a variety of view-points:
“This is an interesting opinion piece that suggests ‘upscaling’ our genomics approaches on non-model organisms, and building ENCODE-like programs to deliver ‘systems biology’ level understanding of a wider range of organisms than those currently studied.” Mark Blaxter
“Proposing a more complete annotation of additional genomes is an interesting idea and well worth a public debate.” Brian Oliver
“There are a number of things about this perspective that don’t sit well with me. First, I am a typical maverick scientist; I run a small lab and prefer to pose my own questions and design my own approaches.” Jim Marden
Our online magazine, Biome, has also published an interview with Jeffrey Boore exploring the need for a neoENCODE project and how it can help existing genome projects.
If you would like to continue the conversation we look forward to receiving your comments! Or join the debate on twitter #neoENCODE.
Elizabeth Moylan & Catherine Rice, BMC Editors.
- Beyond ENCODE – let’s continue the conversation - 22nd July 2014
Comments