
Recent developments in genome-editing technologies have turned the biotech industry's dream of mastering nature into a fast approaching reality. Want to change the color of an ear of corn? All you need to do is edit the relevant gene using the CRISPR/Cas system.
But some traits cannot be edited at the genomic level. The three ears of corn above may have strikingly different appearances, but they are genetically identical – varying only in their epigenomes.
Biotech strategies using genome engineering are impotent when it comes to epigenetically regulated phenotypes. Instead, epigenome-engineering tools are called for. And, as participants heard at May's meeting of the London Epigenomics Club, these prophesized epigenome-engineering tools are now a reality.
How to edit the epigenome
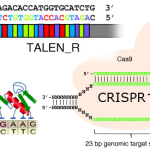
The basis of these systems is that DNA-targeting components of genome-editing tools can be combined with epigenome-modifying enzymes, in order to erase or introduce epigenetic marks. Because the epigenome has evolved to be modifiable in response to changing circumstances, epigenome edits are readily reversible.
Many interesting biological questions are being addressed with epigenome editing: is promoter DNA methylation a cause or a symptom of changes in gene expression? Do different DNA methylation sites act in concert? What are the functional differences between histone modifications? Can DNA be rewound by remodeling nucleosome occupancy?
An alternative editing strategy: small RNAs

So closely intertwined, in fact, that non-coding RNAs can be used as an alternative strategy for editing the epigenome via RNA-directed DNA methylation mechanisms: Bond reported the use of 21 nt small RNAs for initiating de novo DNA methylation at target sites, and Miska hinted that something similar may be possible in nematodes.
How necessary is the epigenome?
DNA methylation, chromatin and non-coding RNAs can all variously be described as components of the epigenome, if we are to set aside semantic arguments. Certainly, they are all mechanisms of both gene regulation and genome defense. But do cells really need so many different flavors of epigenome to regulate their gene content and defend their genomes?
Probably not, thinks Eric Miska. He pointed out that, across the tree of life, different combinations of DNA methylation, chromatin and non-coding RNA systems have arisen and been lost. Take his model organism of choice: the humble nematode, C. elegans.
Genome defense eats itself
The likely ancestral function of DNA methylation and silencing RNAs is genome defense: protection against the mobile genetic elements whose invasion all genomes are vulnerable to.
The most powerful epi/genome editing tool is CRISPR/Cas, which in nature – as opposed to biotech – functions in genome defense.
![]() Curiously, Eugene Koonin and colleagues report in this month's BMC Biology that the origins of CRISPR/Cas itself lies in a mobile genetic element – a class of archaeal self-synthesizing DNA transposons they name 'casposons'.
Curiously, Eugene Koonin and colleagues report in this month's BMC Biology that the origins of CRISPR/Cas itself lies in a mobile genetic element – a class of archaeal self-synthesizing DNA transposons they name 'casposons'.
With epigenome engineering, biotech forms of CRISPR/Cas – derived from tranposons by way of adaptation to genome defense – can be used to defend genomes against the parasitic DNA from which they arose.
Latest posts by Naomi Attar (see all)
- tRFs and the Argonautes: gene silencing from antiquity - 2nd October 2014
- Keeping up with the Jobses: the role of technology in reproducible research - 26th September 2014
- How to disarm a superbug – a story told by forensic genomics - 23rd June 2014
2 Comments