![]() In Greek mythology, the Argonauts are a band of heroes who accompany Jason on his quest to find the Golden Fleece, a garment whose origins likely lie in the use of sheep fleeces as sieves to collect gold flakes from running water.
In Greek mythology, the Argonauts are a band of heroes who accompany Jason on his quest to find the Golden Fleece, a garment whose origins likely lie in the use of sheep fleeces as sieves to collect gold flakes from running water.
In a new paper published in BMC Biology, Anindya Dutta and colleagues mine Argonaute (sic) datasets for biology's very own hidden gold: previously neglected fragments of tRNA molecules, known as tRFs.
Here are seven awesome things you need to know about tRFs:
1) tRNA molecules are routinely degraded by the cell into tRNA halves and smaller fragments (tRFs), which can be created from both the 5' and 3' ends of each tRNA. Some studies have argued that these degradation products are not merely waste, but instead have their own biological functions.
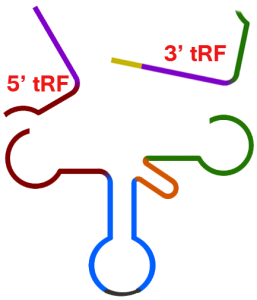
2) Argonaute proteins bind microRNA (miRNA) molecules as part of a process of gene silencing. These interactions are studied using techniques such as CLIP and CLASH.
Anindya Dutta and colleagues have noticed that hidden in CLIP and CLASH datasets is plenty of evidence that tRFs also bind to Argonautes – a fact that has often been overlooked owing to a focus on miRNAs.

3) miRNAs and tRFs share many similarities. Both are small RNAs. As with miRNAs, tRFs (when bound to Argonautes) seem to downregulate, or 'silence', mRNAs containing complementary sequences.
They form similar complexes with Argonautes and target mRNAs, even using the same 'seed' rules for selecting target binding sites. And they are present in the cell at a similar abundance to one another.
Given that the similarities between miRNAs and tRFs are so striking, it is perhaps unsurprising that in at least one case a miRNA reported in the literature turned out to be a tRF.
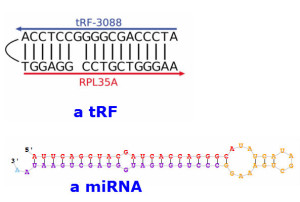
4) But tRFs are not the same as miRNAs. As an obvious difference, tRFs are created from pol III-transcribed tRNAs, whereas miRNAs are the products of pol II-transcribed pre-miRNA molecules. miRNA production requires the DROSHA and DICER1 enzymes, but tRF generation is DROSHA- and DICER1-independent.
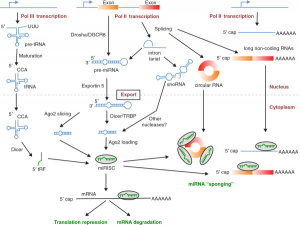
5) Another difference between tRFs and miRNAs is the identity of the Argonaute proteins to which they bind. For the most part, miRNAs form complexes with Argonaute 2 but not Argonautes 1, 3 or 4. For tRFs, Argonaute preference is reversed, with Argonautes 1, 3 and 4 being favored, and Argonaute 2 shunned.

6) Perhaps the most curious difference of all is that, unlike metazoan-and-plant-specific miRNAs, tRFs are ubiquitous.
You can find them everywhere from bacteria, to people, to plankton (a BMC Genomics paper recently spotted tRFs in the diatom Phaeodactylum tricornutum), to plants (as shown in this Biology Direct paper).
tRFs offer one explanation as to why bacteria have Argonaute proteins despite not possessing a miRNA pathway. Jason and the Argonauts is a legend of great antiquity, and we can infer from their ubiquity that tRFs are also very ancient – most likely an ancestral gene-silencing mechanism present in the last universal common ancestor.
7) Dutta and colleagues have constructed a database (maybe they used JSON?) of Argonaute-interacting tRFs, which they hope will launch other curious biologists on their own odysseys of discovery.
But, wait, there's more!
The tendency of tRNAs to break down doesn't stop with tRFs. As stated above, they can degrade into larger pieces, known as tRNA halves, which have also been shown to play a regulatory role in gene expression.
Whereas small tRFs target mRNAs for silencing through interactions with Argonautes, tRNA halves use an altogether different mechanism. Taking advantage of their ability to mimic full-length tRNAs, the halves seek out tRNA-binding sites in ribosomes. Occupation of these sites can stall mRNA translation and so downregulate protein expression.
Even before they are transcribed into RNA, tRNAs can fall apart. That is, tRNA genes are sometimes split. A number of organisms, mostly but not exclusively archaeal species, have split some of the tRNA genes in their genomes into two. In other cases, they are not split, but permuted.
Between tRFs, tRNA halves and fragmented tRNA genes, it seems that tRNAs just don't like to be whole.
Latest posts by Naomi Attar (see all)
- tRFs and the Argonautes: gene silencing from antiquity - 2nd October 2014
- Keeping up with the Jobses: the role of technology in reproducible research - 26th September 2014
- How to disarm a superbug – a story told by forensic genomics - 23rd June 2014
[…] In Greek mythology, the Argonauts are a band of heroes who accompany Jason on his quest to find the Golden Fleece, a garment whose origins likely lie in the use of sheep fleeces as sieves to collect gold flakes from running water. […]
[…] In the years since the first draft of the human genome was released, our understanding of the incredible complexity that lies beyond protein coding regions has grown considerably. Of particular note is the growing family of non-coding RNAs. In a recent study in BMC Biology, Anindya Dutta from the University of Virginia School of Medicine, USA, and colleagues probe one of the newest members of this family, tRNA derived RNA fragments (tRFs). There is still much debate over their biogenesis and function, leading Dutta and colleagues to carry out a meta-analysis of publically available data on tRFs. Through their analysis it was revealed that these fragments are more evolutionarily conserved than their better known contemporaries, the microRNAs, and moreover uncovered that they, like microRNAs, are able to bind Argonaute proteins to form complexes that are involved in gene silencing. Here Dutta explains some of their more surprising results, speculates upon the function of tRFs, and discusses what comes next in the tRF story. More on what makes tRFs such an intriguing class of non-coding RNAs can be found in this BioMed Central blog. […]
[…] tRFs and the Argonautes: gene silencing from antiquity – BioMed Central blog. […]