
It’s been a while since I attended the Annual Meeting of the Ecological Society of America, in person! In the last 10 years, since I became a tenure-track faculty member at Eastern Washington University, a regional comprehensive public university in the US, I mostly had to focus on developing and teaching my classes, which is 80% of my workload. Attending conferences in person is also expensive, starting with registration fees, plane tickets, accommodations and meals. It also takes a toll on a family, missing out on time spent with kids and putting more burden on the spouse staying at home with the kids. All in all, I didn’t have the time and the money to attend in person. I tried attending the conference virtually last summer, but I was not satisfied with the experience. However, this summer I was fortunate enough to be able to attend and present at the Annual Meeting in Los Angeles in person, thanks to an internal grant from my institution!
The Ecological Society of America is the premier organization of all ecologists in the US, and their Annual Meeting is the largest gathering of ecologists, usually with up to 5000 attendees. It lasted for an entire week, from August 4 to August 9, with workshops and invited and special sessions, and contributed talks, and posters and mixers, social events, field trips, career development and training opportunities and of course plenary talks. From Monday afternoon until Thursday evening, there could be up to 25 concurrent sessions going, so you really had to make hard choices on what talk to attend at any given time and peruse the schedule to plan your day. Being a disease ecologist, I mostly attended the Disease and Epidemiology sessions on Wednesday and Thursday.

I made a couple of general observations. For one, mostly the same, roughly 100 people (the disease ecology community) attended the sessions, a good mixture across the academic spectrum, from undergraduate to graduate students to postdocs to junior and senior faculty. This reflected the academic stage of the presenters as well, most of them being PhD students presenting their thesis projects, working in the labs of faculty I was still familiar with, but also postdocs and faculty and even senior faculty presenting. In addition, there were also some speakers working for government agencies and non-profits. Finally, a few of the presenters were undergraduates and even high schoolers. There was a great variety of approaches presented from pure mathematical modeling to statistics to field observations and molecular biology, and even some social science. Topics ranged widely from human and animal pathogens (including hantavirus, influenza, dengue, SARS-CoV-2, West Nile virus and Pd), but also plant microbiome and pathogens, such as BYDV in grasses and the fungal pathogen Ceratocystis lukuohia, causing Rapid ʻŌhiʻa Death in Hawaii. Climate change and the validity of the transmission-virulence trade-off was one of the common themes across many of the talks I observed.
One of the greatest benefits of attending the conference for myself was learning about a lot of new developments that I will be able to share with students in my classes in the years to come. For example, I wasn’t aware of the great variety of different databases that are now available to be mined and used by students anywhere, including at EWU, about pathogens and their hosts and their characteristics. While I have heard about the USAID PREDICT database before, through the special session organized by the Verena Institute, I learned about the ZOVER database on zoonotic viruses, the VIRION database on vertebrate-virus networks, and the PanTHERIA dataset on mammals, that they were all using to try to predict and prevent the next pandemic. In the interesting discussion that followed, the presenters identified the lack of long-term datasets on the pathogen exposure and infection status of the same individuals over long time as one of the challenges to their work (although I would argue that veterinarians, farmers, zookeepers and animal shelter and rescue operators would be excellent sources of those observations for domestic animals and livestock at large). I also learned separately that there is a Lyme disease vaccine being tested to protect mice from getting infected, similar to the oral rabies vaccine. I received excellent feedback on my presentation on the research of my graduate student Sarah Flores on the seasonal prevalence of a trophically transmitted parasite in an invasive fish at a National Wildlife Refuge. I got some cool and useful ideas on the future direction of research in my lab, from how to estimate West Nile virus risk by combining many different sources of information, to potential methods to control ticks in wildland settings. Finally, I was able to catch up with some old friends and colleagues and make some new contacts, which might lead to new opportunities and funding for myself and my students.
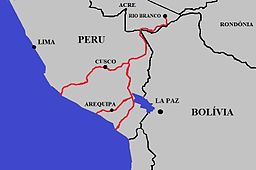
I can’t possibly do justice to the 44 talks I watched and their authors in a single blog post. Instead, I will just highlight three labs that made a huge impression on me subjectively. The one that blew me away the most was Erin Mordecai from Stanford talking about the effects of land-use land-cover change on the distribution and prevalence of vector-borne diseases in South America. First, she shared her findings linking the construction of the Inter-oceanic highway in Peru to increases in dengue importation and transmission, while correcting for many covariates, using the “difference in differencies” approach of causal inference. She also used the same methodology, which she advocated others in audience to use, to link the increase in gold mining in Brazil during the Bolsonaro administration with increases in malaria cases, especially among the Yanomami people. She convinced me enough to look into the methodology, used by social scientists, and understand that it’s different and potentially superior to ordinary multiple regression or partial regression techniques. Next, I was amazed by the variety of research done in Joe Hoyt’s lab at Virginia Tech. I will here focus on his study of SARS-Cov-2 in wildlife, both kept in rehabilitation centers and actively captured. While he and his students found the virus in a large variety of wildlife (e.g. deer mice, possums, racoons), with seroprevalence increasing with increasing human presence, the viral strains detected were representative of those circulating in humans, not suggesting new variants lurking in wildlife, ready to jump back to us.

Finally, I’d like to highlight Ariel Greiner‘s talk, a postdoc of Matt Ferrari at Penn State. She modeled the best surveillance methods to detect and control Foot-and-Mouth Disease in cattle with limited resources, based on a unique dataset from the Republic of Turkiye, with information on the distance and connection between cattle farms in the country and the number of outbreaks at each farm between 2007-2012, using time-variant networks. She found that focusing on a small portion (say 20%) of the most highly connected farms was equally superior to focusing on the farms closest to prior outbreaks, or farms that were most connected to the farms with prior outbreaks, compared to random sampling. What I find amazing in these results is that it can be used solely based on knowing the degree of connections between farms, without any disease data, hence it could be used to set up a surveillance system 2-4 times more efficient than random sampling in the absence or in anticipation of the introduction of an outbreak of a new disease. I do hope that regulatory agencies, such as the USDA, will take note of her research and implement it in their practices!
Above, I was obviously only able to scratch the surface of the experience I had at ESA this year, which was also a tiny portion of everything it had to offer. If interested, you can see the entire schedule and browse all the abstracts of the conference here. I do hope that I conveyed how much fun I had, and how grateful I am to have been able to attend and present. I very much hope that I will be able to attend again and can wholeheartedly recommend all of you to do the same! Looking forward to meeting up with all of you in Baltimore next year!

Comments