
Parasites have long been known to interact greatly with their host organisms, and are even able to significantly alter the fitness of their host’s – a process thought to increase the parasites chances of successfully completing its life cycle.
For example, rodents infected with one of the world’s most common parasites, Toxoplasma gondii, has been well demonstrated to increase risky behaviour towards feline predators (the definitive hosts of T. gondii). This ingenious parasite has also been shown to cause increased aggressive and impulsive behaviours in humans, and curiously, possibly even increased entrepreneurial activity.
A great exemplar of both host-parasite interactions and how parasites can alter their host’s behaviours for their own benefit is found in the infection of the three-spined stickleback (Gasterosteus aculeatus) by Schistocephalus solidus.
Infection-induced host behavioural changes can range from slight alterations in pre-existing traits (such as the increases mentioned above), to the formation of entirely novel behaviours.
S. solidus is a parasitic tapeworm (cestode) that utilises three host organisms to complete its life cycle; a cyclopoid copepod (small crustacean), the three-spined stickleback (teleost fish), and fish-eating birds. The two former hosts are intermediate species, and the fish-eating water birds act as the definitive host’s for S. solidus, providing a suitable environment for the parasites hermaphroditic reproductive process.
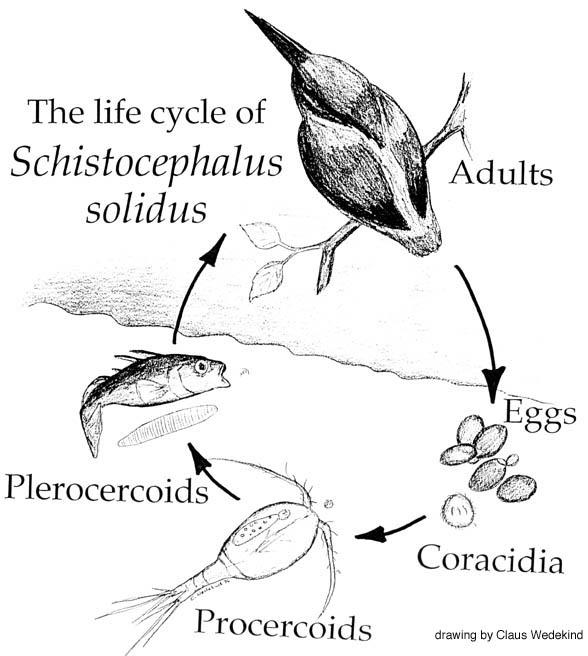
As described in two previous blogs, S. solidus has already been demonstrated to alter the behaviours of its copepod and stickleback intermediate hosts. In an infected stickleback host, changes to physiology (e.g. increased O2 consumption and decreased energy reserves) have been shown, as well as alterations to shoaling behaviour, loss of anti-predator responses, and increased proximity to the water’s surface.
Even though the effects of parasites on infected hosts have been well observed, there is comparatively little information on the molecular pathways involved in the drastic behavioural changes seen, including in the S. solidus-stickleback system. Based on the physiological and behavioural alterations mentioned above, it can be assumed that the mechanisms that induce these alterations must include interconnected structures and processes such as neural circuits, neuroendocrine regulation, gene expression changes, and epigenetic regulation.

In a recent publication, Lucie Grecias and colleagues investigated the mechanistic basis of interactions between host and parasite in the context of parasite-induced behavioural changes in the host.
To achieve this, the genome-wide expression of genes in the brains of sticklebacks were investigated using RNA-seq.
- Firstly, the researchers analysed transcriptomes of infected sticklebacks. They predicted that sticklebacks would show changes in gene expression related to the behavioural, physiological and host response to infection changes they exhibit.
- Secondly, they used exposed sticklebacks in which tapeworms did not develop. They predicted that exposed fish would share a pattern of gene expression with infected fish, due to the common expression of genes related to host responses to infection.
- And lastly, some sticklebacks were treated with an SSRI (Selective Serotonin Reuptake Inhibitor-treated), as this drug is known to cause similar behavioural changes to those induced by solidus-infection. The researchers predicted that if the SSRI affects the same molecular pathways as S. solidus does, then substantial overlap would be detected when comparing gene expression in SSRI-treated and infected sticklebacks.
Their investigation was thus comprised of four experimental groups: healthy-control sticklebacks, S. solidus-infected sticklebacks, parasite exposed (but not infected) sticklebacks, and SSRI-treated sticklebacks.
Altered molecular pathways in the brain following S. solidus infection
The researchers discovered that 105 genes were differentially expressed between infected and healthy sticklebacks. Forty-five of these genes were differentially expressed in only the comparison of infected and healthy-control fish, and are thus considered as a genomic signature of infection.
Of those 45 genes, several genes coding for molecules involved in the inositol pathway were found to be differentially regulated in brains of infected fish. One of these genes, IMPA 1, codes for IMPase 1– an enzyme central to inositol metabolism. IMPase 1 was upregulated and expressed more highly in brains of infected sticklebacks compared to exposed, SSRI-treated and healthy-control fish.
In humans, altered inositol metabolism has been implicated in a number of neurological diseases, such as manic symptoms seen in bipolar patients. In these patients, lithium chloride is used to lessen behavioural symptoms of the disease by blocking IMPase 1 activity. Therefore, the researchers tested to see if they could ‘rescue’ two altered behaviours in S. solidus-infected fish (a tendency to swim close to the surface, and a lack of response to predator attacks) by utilising lithium in the same way.
Their results showed that the treatment made infected sticklebacks behave (in part) more like healthy fish, therefore implying that alterations in the inositol pathway could be a direct cause of the dramatic behavioural alterations observed in the S. solidus-stickleback model system.
Overlap of exposed and infected fish transcriptomes
Results of this analysis showed infected and exposed sticklebacks did not have similar brain expression profiles, contrary to their prediction. Intriguingly though, gene expression in brains of exposed fish were also different from healthy fish, therefore producing an ‘exposed’ genomic signature. It seems that even exposure to S. solidus has significant effects on stickleback brain chemistry.
Overlap of SSRI-treated and infected fish transcriptomes
The analysis of shared gene expression in brains of SSRI-treated and infected sticklebacks revealed only four common genes, which are involved in cellular organisation and transport. This small overlap may be explained by SSRIs being specifically designed to act on a unique target, whereas S. solidus affects numerous traits, as well as behaviours. Also, it does not necessarily suggest that SSRIs and S. solidus target different molecular pathways, only that the observed similarities are masked by multiple molecular differences.
Conclusion and future prospects
This work by Lucie Grecias and colleagues aimed to determine the brain gene expression profiles of parasite-infected sticklebacks to identify genomic signatures differing from healthy-control sticklebacks. Their results enabled the prediction that S. solidus directly manipulates its stickleback host via the inositol pathway. Although, as the authors note, tests are needed to discover if the altered inositol pathway is a side-effect of a host response to infection, or if it occurs as a direct result of manipulation by the parasite. Thus, the next natural step for the team is to determine if S. solidus acts to alter its hosts behaviour by specifically manipulating IMPase 1 and the inositol pathway, and if sticklebacks with experimentally altered pathways experience the same altered behaviours that increase their probability of predation by the definitive avian host.

Comments