
Chinese, Japanese, Korean, what is the difference? It is usually difficult to tell which of the three East Asian groups a person comes from just by looking at their appearance. Indeed, these three influential ethnic groups, i.e., Han Chinese, Japanese, and Korean share many similarities in appearance, language and culture.
Such similarities are also reflected in our genetic data. The genetic difference between any of the three groups is less than 1% of their total genetic diversity, which is much smaller than that between any of the groups and a European population (~10%). Accordingly, the three groups separated from each other from their recent common ancestor only 3,000 ~ 3,600 years ago, roughly corresponding to the Shang Dynasty in Chinese history.
These estimations based on genomic data indicate Han Chinese, Japanese and Korean people are genetically closely-related and derived their ancestry from a common gene pool.
On the other hand genome-wide variation data can largely distinguish Han Chinese, Japanese and Korean individuals without much ambiguity (see the image below).
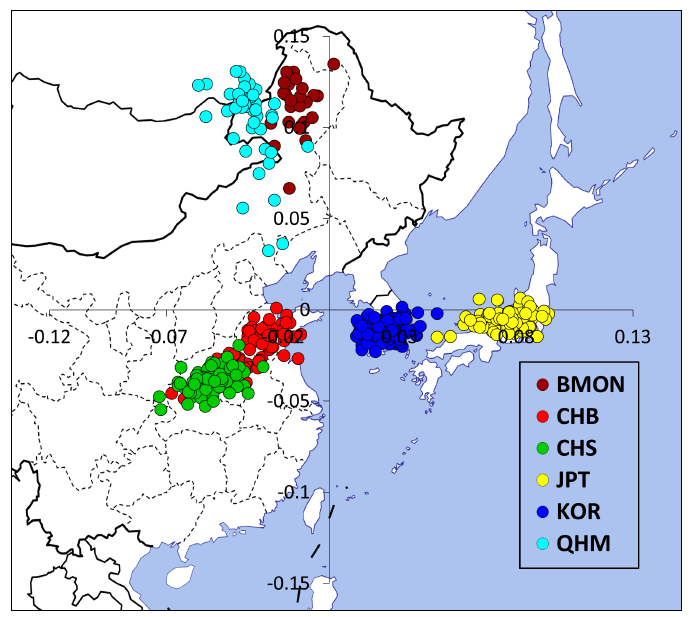
Since the population diverged, the present-day Han Chinese, Japanese and Korean populations have built their own gene pools and formed distinct genetic makeups. This means that individual ethnicity of the three East Asian groups is distinguishable in genetics if personal genome data are available. In general, genetic differences between Japanese and Han Chinese are larger than that between Korean and Han Chinese.
In general, genetic differences between Japanese and Han Chinese are larger than that between Korean and Han Chinese.
The genetic distinctions among the three East Asian groups initially resulted from population divergence due to pre-historical or historical migrations. Subsequently, different geographical locations where the three populations are living, the mainland of China, the Korean Peninsula and the Japanese archipelago, respectively, apparently facilitated population differentiation.
Apart from the overall genome-wide differentiation, genes showing considerable differences have also been identified among the three groups. For example, several highly differentiated variants are enriched in the CD46 gene which is located on chromosome 1. It’s thought that this gene might be associated with human adaptation to pathogens in different local regions. In addition, the protein encoded by this gene may be involved in the fusion of the spermatozoa with the oocyte during fertilization and thus might be related to reproductive traits, although further studies are necessary to validate these signals and interpretations.
This study does not fully address the initial settlement of people in these three countries; further research is needed to disentangle the complex history of the three most influential ethnic groups in East Asia. Genetic basis of distinct phenotypic variations among the three ethnic groups also remains to be resolved by future studies.
11 Comments