With all the fuss around how much CRISPR/Cas-based editing (or as a matter of fact any genome editing technology) can and cannot do, it is easy to forget about one thing: all these wonderful techniques, so simple that anyone can use them, achieve primarily one goal. They cut DNA where we want them.
Which is of course an amazing ability, but only until you realize that once the DNA is cut that, to be of any use, it should be somehow put together again. And, as it turns out, none of how this happens in cells has anything to do with us, experimenters.
Cellular repair team
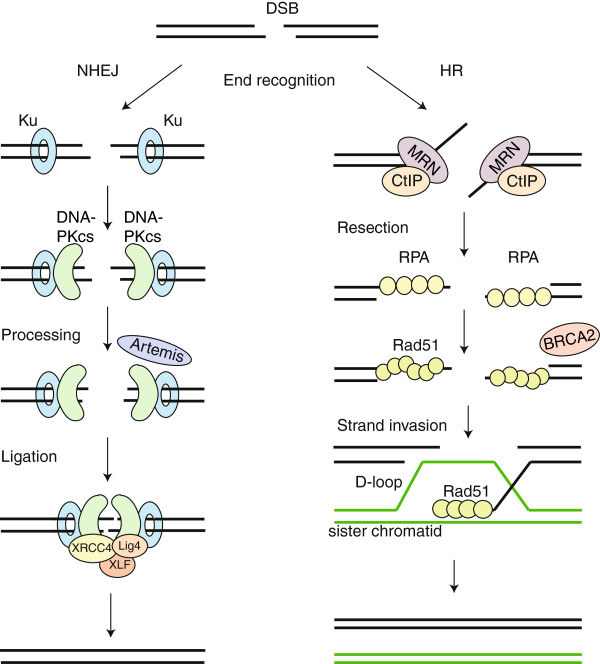
There are two main mechanisms that cells can use to repair damage in DNA caused by genome editing tools: non-homologous end joining (NHEJ) and homology-directed repair (HR). During NHEJ, the DNA is repaired by a direct ligation of cut ends, without the use of repair template of any sort. Depending on the quality of the cuts – that is to say, how compatible are the ends of the DNA strands – the enzymatic mechanisms in place can trim the DNA further or make up some missing sequences.
Since these changes are made without relating to the actual sequence of this region in any way, the end result may be quite different to the original – NHEJ often leads to uncontrollable insertions or deletions.
Now, if our goal is just a disruption of a gene, then this probably isn’t a problem. However, the most interesting applications of genome editing technologies (from the human health point of view) are those in therapy. For example, Duchenne muscular dystrophy is caused by mutations in the dystrophin gene – their accurate repair is considered as a viable treatment for this disease.
Here though the NHEJ is not enough. This is where the other repair mechanism comes in. Homology-directed repair, as the name suggests, uses homologous templates to repair DNA breaks. The mechanism of repair is much more complicated than for NHEJ, and also requires an introduction of a template sequence. While it allows for much more precise DNA repair, it suffers from some problems that hugely affect how useful it is in genome editing – and so how useful these editing approaches may be for therapeutic purposes.
The type of repair mechanism used by the cell depends to a large degree on the complexity of the break. In the majority of cases cells use the ‘quick-and-dirty’ NHEJ. As a matter of fact, NHEJ is so much simpler and faster than HR that it often completely drowns the homology-directed response.
This means then during genome editing experiments the repair efficiency achieved with NHEJ can reach anywhere between 20 and 60% (and continuous developments help improve these numbers all the time). Homology-directed repair efficiency at the same time usually trails in the area of fractions of percent.
For advice on what is best for HR, ask your local chemist
Earlier this year, however, two studies in Nature Biotechnology, one from Hidde Ploegh and colleagues and the other from Ralf Kühn and colleagues, showed how homology-directed repair efficiency can be improved (up to 19-fold and up to 8-fold, respectively) simply by inhibiting some of the key NHEJ players. By preventing the cells from using NHEJ, the homology-directed repair is promoted.
Last week, another piece was added to this growing body of work on improving HR efficiencies – Jerry Pelletier and colleagues published an article in Genome Medicine describing the effect of inhibition of DNA-dependent protein kinases involved in NHEJ on the homology-directed repair in CRISPR/Cas9 experiments.
The authors tested two small molecule DNA-PKcs inhibitors, poetically called NU7441 and KU-0060648. Perhaps unsurprisingly, given the known function of these molecules as well as the enzymes which they affect, the frequency of NHEJ in the tested cells dropped. At the same time they observed an increase in homology-directed repair. These molecules give a HR efficiency improvement at the levels similar to tools described in previous papers, thus expanding our repertoire of available HR promotion means .
An interesting aspect of the Pelletier and colleagues’ study is the type of molecules used to stimulate homology-directed repair. While the previous studies used gene silencing approaches, the authors here use simple molecules that can be bought pretty much off the shelf, making this substantial improvement in the genome editing technology achievable to many laboratories less experienced in molecular level manipulations.
And so it goes
If you enjoyed reading about the Pelletier and colleagues’ article, please do look out for Genome Biology special issue on genome editing – this will be published in November 2015, and while the submission deadline has passed now, we are still considering late submissions.
Comments