On 28 August 2014, the magazine Science published an important article: the results of sequencing of 99 Ebola genomes. Since the confirmation of several cases of the disease in Sierra Leone in May earlier that year, the scientists collaborating with local authorities started to send out samples to the Broad Institute, where these were further sequenced and analyzed. The study let us understand the origins of the disease, as well as how it evolved as the epidemic spread.
The study was of no small importance, scientifically and medically, and an example of overcoming numerous challenges. During the work on the Ebola genomes, the scientists developed new methods that should make similar studies in the future easier, faster and more achievable (one such computational method was published in Genome Biology‘s special issue on infectious diseases). We can only imagine the miles of red tape that must have been cut and the logistics and cost of the transport of the (presumably biologically hazardous) patient samples out of Africa. And as we today also know, the results came at a great personal cost to many people.
And yet, the study still arrived too late for over 1,500 patients who were by then already dead (another 8,500 would follow in the coming months). Today, we can speculate: if the sequencing technology was readily available to the doctors of Sierra Leone in the early days of epidemics, is it possible that the spread of the disease could have been contained quicker?
It is, of course, rather impossible to answer this question; and to say that a matter of access to technology could turn the tides in Africa much earlier would be a cruel and unfair assessment of an extremely difficult situation that doctors, scientists and local authorities faced. However, once we separate complicated and emotion-laden circumstances of the recent Ebola outbreak from an abstract hypothetical epidemic scenario, we cannot but stop to wonder: is the on-site genomics surveillance the future of outbreak control? I mean, it must be, mustn’t it?
MinION helps with Salmonella
The easy solution for this would be to have a sequencer in every hospital on Earth. And then, when we stop dreaming, we can ask – is there a more realistic alternative? The answer is ‘yes’ and comes with the latest development in sequencing technologies, nanopore sequencing.
The story goes back to the early summer of 2014, and to another outbreak. This one happened in the United Kingdom, and was only barely reported in the media. But what came out of it may – and likely will – have huge implications for all large epidemics we’ll face in the future.
In June, a Salmonella outbreak at Heartlands Hospital in Birmingham led to the closure of eight wards. The outbreak was sudden, and proved to be an excellent testing ground for a new technology championed by an Oxford-based company, Oxford Nanopore Technologies, which is a manufacturer of the first ever miniature portable DNA sequencer. A group of researchers from Birmingham University led by Peter Hawkey and Nick Loman, set out to identify the origins of epidemics using the MinION device. The results are now published in Genome Biology.
The study demonstrated (at least) two very impressive aspects of the ONT’s device. First, it is the first ever application of a portable sequencer in a clinical setting. While the results obtained with MinION in Birmingham were confirmed with a more established Illumina technology (MiSeq), they are a proof that a device can be used in the field where other larger, more expensive sequencing machines are simply not available.
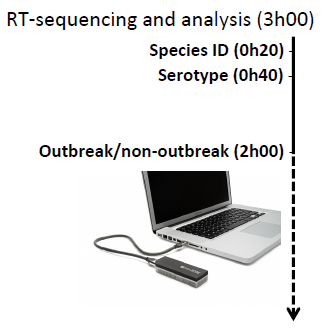
The other aspect was a sheer speed of obtaining meaningful and actionable results. Beyond the sequencing library preparation (which will be a limiting step for any sequencing technology), the sequencing itself, and the data analysis, can be indeed very fast: the researchers were able to identify Salmonella enterica as a responsible species within under half an hour! Within another 50 minutes they knew the serotype to by Enteritidis, and after another 100 or so minutes they were able to say if the strain they observed belongs to an outbreak cluster. In other words, in 3 hours from placing the sample in a sequencer, they knew where they stood with an outbreak.
Translating theory into practice
Now that we know that these machines work as well as they do, it is high time to put them to a proper test: to see if it is possible to take a portable sequencer to a remote location and use it effectively in a way that would inform the local public health responses.
This is democratization of sequencing.
Joshua Quick, University of Birmingham
And this is exactly what is happening right now. Earlier this month, Nature’s News section reported on a trip that Joshua Quick, the first author of the Genome Biology article, made to Guinea in April this year. Quick himself reported on the trip at more length during London Calling, Oxford Nanopore Technologies’ workshop that took place in London mid-May.
Briefly, Quick packed two small suitcases full of essentials: MinION sequencers (of course), a PCR machine, consumables and reagents. And then flew to Guinea to meet an empty room in one of the country’s hospitals where he set up a lab and for the next 12 days sequenced samples from 14 Ebola patients – in one case getting the results within 48 hours from the sample collection.

But this latest project from Nick Loman’s lab is not the only one that is putting MinION devices through the mill: during London Calling two other speakers, Miles Carroll from Public Health England and Thomas Hoenen from NIH, also reported on projects looking at epidemiological surveillance of Ebola, while University of Tokyo’s Yutaka Suzuki described similar efforts with Dengue fever in Indonesia.
Quo vadis, nanopore-seq?
During London Calling Mick Watson of Edinburgh, another nanopore-sequencing enthusiast, proclaimed
And it is, but it is worth remembering about several caveats that are still in place.
Like any emerging technology, nanopore-sequencing offered by ONT, while good, can certainly still be better. The improvements should be expected in the technology itself, for instance, the dreaded chemistry of the membrane, or the dreaded chemistry of the nanopore enzyme. We should also expect – and as a matter of fact we have already seen ONT making good on the promise (and premise) earlier this month – developments of auxiliary devices: liquid-handling systems for easier, faster sample prep, the improvements of the analytical software and so on.
One major obstacle that may affect the utility of the portable sequences is access to Internet, since a lot of the MinION analysis happens in the cloud. Quick (now famously) used 17 SIM cards for his 14 patients. It may then appear that the most costly part of remote epidemics surveillance won’t be sequencing, but data transfer. Interestingly though, this issue may be resolved by another amazing technological project: Google’s Project Loon.
Oxford Nanopore Technologies seems to have made a major leap in sequencing technologies: not only how it is done, but most importantly, what it can be used for. Their interesting marketing strategy made them a frontrunner of the fourth generation sequencing, but we should impatiently await others to promptly follow suit. Because it is increasingly clear that nanopores are the future of genomics.
Comments