Identifying the most appropriate therapies from cancer genome data is a major challenge in personalized cancer medicine. Physicians and researchers are faced with long lists of tumor-specific genomic variants where most variants are clinically “unactionable”- meaning either their biological role is unknown or is irrelevant for tumor biology. In addition, the current list of known cancer driver genes has clinical limitations since genomic alterations in these genes may be essential for tumor development, tumor cell growth and survival but the same genes may not be targetable by current therapies.
This scenario demands novel bioinformatics methods to interpret genomic alterations, evaluate their clinical relevance and drug feasibility, and provide a prioritized evidence-based list of tailored anticancer therapies to facilitate clinical decision making.
Introducing PanDrugs
We have developed PanDrugs, a novel data-driven method to evaluate cancer patients’ genomic profiles, according to their biological and clinical relevance and opportunities for therapeutic intervention. The methodology scans PanDrugs database (PanDrugsdb) and return prioritized candidate drugs and targetable genes estimated from a list of genomic variants (or genes) provided by a user.
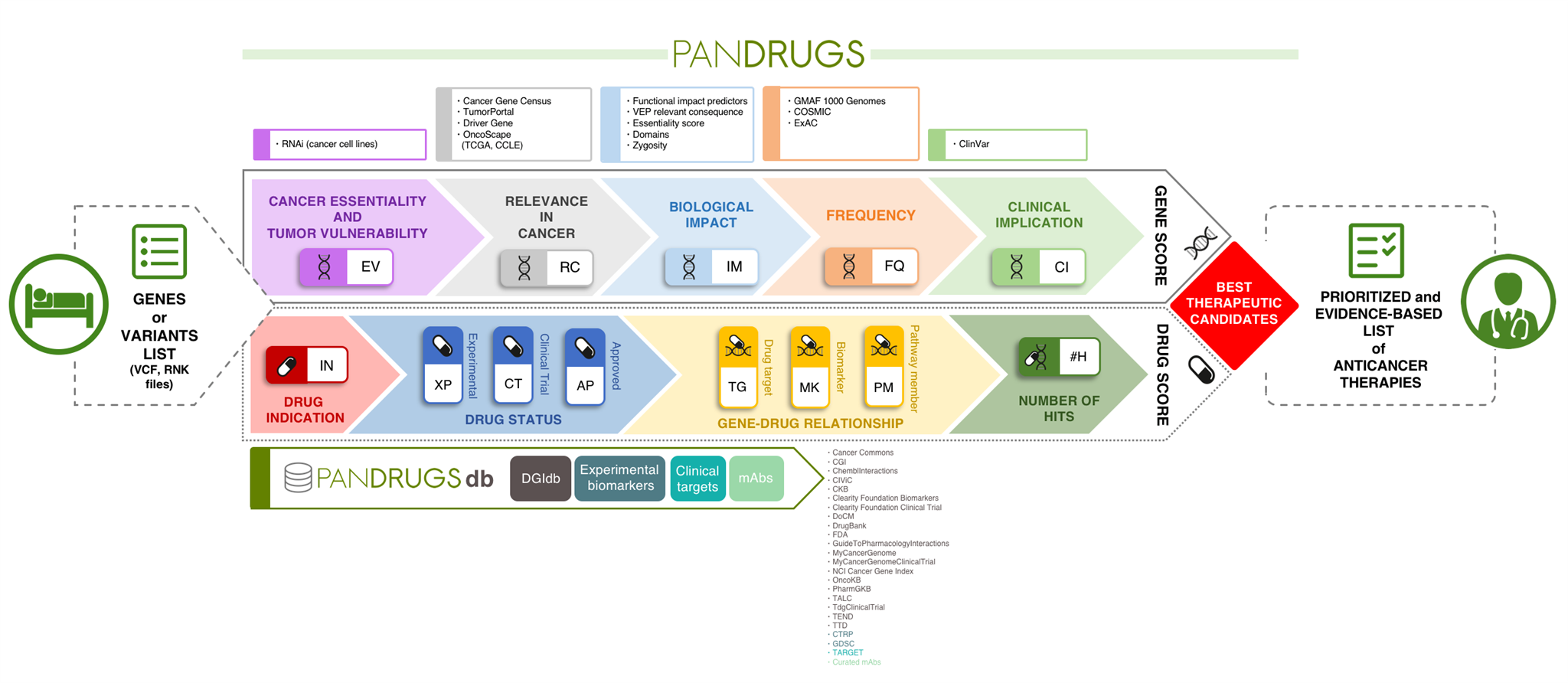
PanDrugs operates by incorporating two scores, which integrate a variety of clinical, biological and pharmacological sources to suggest tailored anticancer therapies. The Gene Score (GScore) is based on the level of evidence supporting gene clinical implication and its biological relevance in cancer and the Drug Score (DScore) estimates drug response and treatment suitability.
Another unique feature compared with current tools is the broadening of the search space to provide therapeutic options. In other words, PanDrugs suggests treatments for direct targets (e.g. genes that contribute to disease phenotype and can be directly targeted by an available drug) and biomarkers (e.g. genes that can be predictive of a drug response but the protein product is not the drug target itself).
However, PanDrugs also integrates a systems biology knowledge-based layer that automatically inspects biological circuits expanding cancer candidate therapies beyond cancer-related gene lists to the whole druggable pathway. This novel “pathway member” strategy extends the treatment opportunities of cancer patients and opens new avenues for personalized medicine. In the article, we apply this strategy to show how prostate, breast and colorectal cancer patients without druggable cancer driver altered genes would be benefited by treatments used in clinical practice.
The PanDrugs database itself represents a valuable publically-accessible resource. This database is the largest public repository of drug-target associations available from well-known targeted therapies to preclinical drugs. Current version of PanDrugsdb integrates data from 24 primary sources and supports >56,000 drug-target associations. Both PanDrugs and PanDrugsdb are open-source and fully available at https://www.pandrugs.org.
Translational applications
PanDrugs supports clinical decision making by providing a way to study molecular profiles in cancer patients, and it can be easily implemented with others variant analysis software in healthcare institutions. PanDrugs is being extensively used to analyze patients´ sequencing data generated in our institution and propose treatments – an example of which is described in our Genome Medicine article.
In addition, PanDrugs has been systematically applied on a cohort of 7,069 patients from the TCGA project that correspond to 20 different tumor types. Full results may be interactively accessed at the PanDrugs website.
PanDrugs perspectives
Personalized cancer treatment is still taking its first steps. Before it can become the norm, hospitals need quick, reproducible, approachable and low cost computational methods to detect altered genes, to interpret the biological and clinical impact of such alterations, to establish therapeutic guidance based on the patient’s genomic profile and to achieve knowledge-driven clinical assessment though health-record data mining.
PanDrugs is the first method to systematically infer novel targeted treatments following a rational framework supported by multi-gene markers, molecular pathway context and pharmacological evidence.
Although PanDrugs offers a valuable methodology for in silico prescription, our method is limited by the lack of large longitudinal precision medicine studies with accessible medical records. Such studies are crucial to assess and validate drug proposals and improve in silico prescription algorithms that consider additional factors such as mode of drug administration, combinatorial therapies, drug repositioning and side effects. Further efforts are also required to improve cancer treatment by developing more effective drugs and anticipating drug resistance.
Nowadays, there are significant challenges that need to be addressed in order for precision medicine to be broadly established in the hospital environment; however, PanDrugs contributes one of the missing links between the potential of biomarkers as predictive tools for treatment outcome and their actual use in clinical settings.
The authors want to thank PanDrugs users for sending valuable suggestions to improve the method. They also encourage cancer researchers and physicians to try PanDrugs by themselves and send feedback to bring it closer to the daily clinical practice.
2 Comments