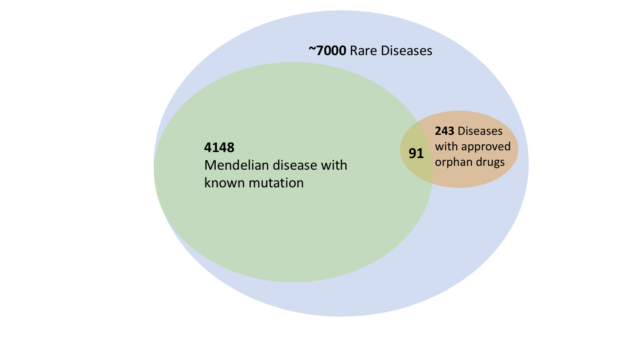
Rare diseases are more common than you might think. Each individual rare disease afflicts fewer than 1 in 2000 people. But collectively, these diseases impact more than 7% of the total US population. This seeming paradox is driven by the abundance of rare diseases: more than 7,000 in total, according to the US FDA.
We are not doing enough to help people suffering from rare diseases. In a recent study, we found that fewer than 4% of rare diseases had drugs approved by the US FDA in 2015. There is limited research funding for the study of these diseases, and little market opportunity to encourage private drug companies. Most patients with rare diseases go through a lengthy process to obtain their diagnosis. It is frustrating that doctors often cannot offer any specific treatment for them.
Can we learn something from the 4% of rare diseases that have a treatment? Patterns across these success stories could highlight a way forward for the diseases without treatment. Decades of biological and patient data have accumulated across public databases, but the scale of these data make curation a challenge. Researchers are not familiar with the thousands of other diseases outside their areas of expertise.
Computational methods such as natural language processing (NLP) offer powerful tools for processing biological data at scale. In our recent work, we created a pipeline that searches for patterns across rare diseases by mining the text descriptions of rare disease in the Online Mendelian Inheritance in Man (OMIM) database and the US FDA orphan drug list. This pipeline connects rare diseases with properties of interest by searching for relevant terms in their text descriptions (e.g. “over activation”). In the aggregate, these properties then reveal many clinically relevant patterns.
Our work highlights promising directions for the treatment of rare diseases. We find that a rare disease caused by over-activated genes is 50% more likely to have a drug in the FDA approval pipeline because 80% of known drugs are inhibitors— chemicals that interrupt biological processes. It is easier to develop drugs to inhibit a gene product, compared to drugs to rescue loss-of-function genes. The field can consider prioritizing funding and efforts for such diseases.
Among the thousands of rare genetic diseases in our analysis, we have identified 34 particularly promising candidates for drug development. Some of these diseases are already receiving attention from the field.
For example, one of our candidates, Achondroplasia, is known to be caused by over-activation of growth inhibition receptor (FGFR3), and currently has drugs under development. However, other candidate diseases have been studied less, such as Parkinson’s Disease subtype 4 and 8. These diseases may particularly benefit from new research attention.
Computational tools allow us to review an enormous amount of biomedical data to discover new knowledge. This knowledge in turn can bring hope to underserved patient populations. The top rare disease candidates from this study are promising new targets for drug development.
Comments