
More and more, ‘omics’ research (e.g., genomics, metabolomics, proteomics) is examining the role of the environment on human health. In a recent symposium held at Yale School of Public Health, the evolving field of the exposome was explored.
The exposome aims to assess the totality of lifetime exposures (e.g., environment, diet, behavior and internal processing from the microbiome, hormones, metabolism etc.) and their combined effect on human health. This concept relies on the fact that genes alone account for the minority of disease etiology for many important illnesses, such as cancer. Research has shown that environmental and lifestyle influences play a critical role in the development of these illnesses.
Genomics and the development of genome-wide association studies (GWAS) have unlocked the ability to explain heritability for phenotypes of interest, such as the risk of developing cancer and other diseases. However, additional technologies are needed to assess the intricate relationships between genes and environment in order to fully understand the mechanisms of disease and identify pathways for intervention and therapeutic design.
In our recently published article in the journal Human Genomics, we discuss how metabolomics, the study of all the small molecular weight metabolites in a system, is an essential tool to examine the exposome, and how major technological advancements have made it possible to use metabolomics to study the exposome.
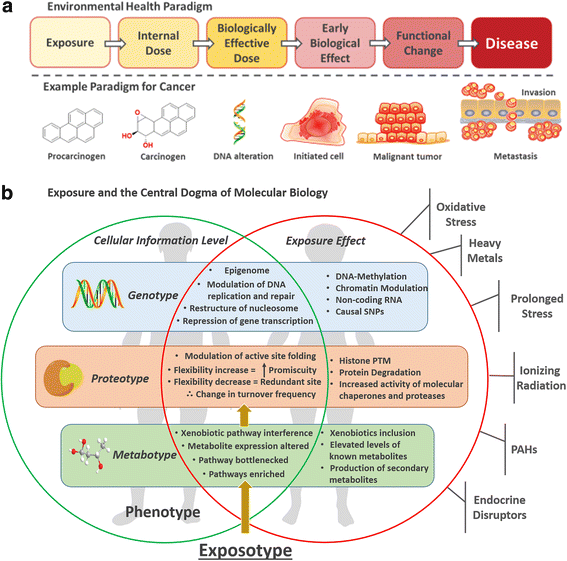
Metabolomics can be used to create a holistic snapshot of the effects of just one incident of environmental exposure on all biological systems within the body. This can then be used to generate an expressed phenotype of exposures: the exposotype. The exposotype also takes into account all exposures experienced including those from lifestyle choices and the current physiological state of the individual.
The combination of metabolomics and GWAS research has the potential to drive significant advances in precision medicine approaches and pharmacogenomics…
Metabolomics can also be used to develop metabolic biomarkers of exposure to environmental factors. This can then be used to build models that can identify associated genetic and enzymatic changes that allows researchers to further understand the molecular mechanisms that underpin the route and consequences of the exposure on the host. This ultimately allows researchers to identify resulting exposotype from the incident of environmental exposure. The combination of metabolomics and GWAS research has the potential to drive significant advances in precision medicine approaches and pharmacogenomics, both of which have been inefficient up to now.
Metabolomics is currently being incorporated into numerous population-level exposure studies to identify gene-environment interactions and forecast disease prevalence by using pre-diagnostic metabolic signatures (collections of metabolites that change prior to disease onset) and genetic risk data. In the future, the combination of metabolomics with genomics offers one tool that may prove helpful towards custom-tailoring precision medicine by identifying the key factors that are related to non-responders and responders of specific treatments. Further advancements are needed in the routine application of appropriate statistical tools as well as the adoption and promotion of transparent and reproducible research practices. This will enable for the science of metabolomics to be more effectively utilized in the identification of individuals at risk for certain diseases and their best treatment options based on how responsive or non-responsive they are likely to be.
While there are challenges involved in metabolomics analysis (such as metabolite identification, analytical bias, controlling for multiple levels of confounders, and reproducibility in large-scale population consortia), these challenges can be overcome through the incorporation of decades worth of research and lessons learned in genetic epidemiology related to study design and statistical analyses. In addition, the growing desire for and recognition of the need for improved transparency, reproducibility and replication has driven efforts in this area in the research community. Current examples of this in the world of metabolomics and exposotype research include databases for exposure metabolites, and new chemometric and bioinformatic tools for data analysis, and integration of multiple types of systems-levels (or “-omics”) data with more on the way.
The exposotype allows greater understanding of the factors that influence human health which include genetic makeup, environmental exposures, lifestyle choices and the interactions between them. Through a general community momentum towards open research practices, improvements in reproducibility, and data sharing, GWAS and metabolomics research examining the exposotype and the exposome, will improve the outlook for many suffering from deadly diseases through the development and application of precision medicine and pre-diagnosis risk assessment.
- Metabolomics and the Exposotype: Linking genes and environment to human health - 9th February 2018
Comments