With a justifiable claim to being the world’s first pseudoenzyme conference, a meeting held recently in Liverpool focused on research into a group of proteins that look like enzymes but aren’t.
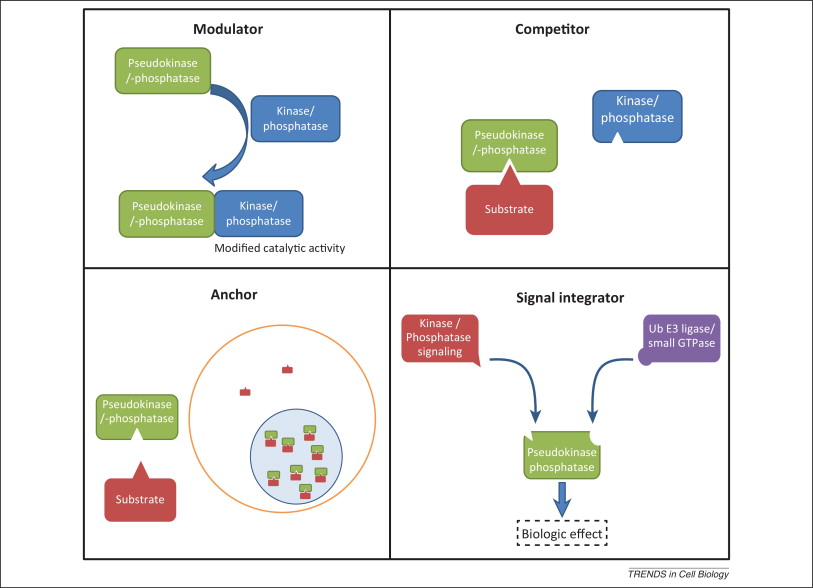
Pseudoenzymes are defined as proteins that are structurally similar to active enzymes but which lack catalytic activity because of mutations to critical amino acids. They are catalytically inert, but nonetheless functional. Hesso Farhan summarised nicely in his talk how pseudoenzymes can retain signaling functions. First, they can anchor other proteins where they are needed. Second, they can modulate the function of other signalling pathway components through, for example, allosteric effects. Third, they can compete with active paralogs for substrate, but fail to catalyze said substrate, reducing the efficiency of the process. Fourth, they can link separate members of signaling pathways together as scaffolds.
The conference provided excellent examples of each of these functions. Hesso Farhan showed how pseudoenzymes can have a substrate-competitive mechanism with the example of STYX’s involvement with FBXW7 and the relationship between the two in breast cancer.
An example of a pseudoenzyme capable of performing two of these functions at once, in the regulation of asymmetric cell division in Caulobacter crescentus, was provided by Seth Childers’ work on DivL, which both anchors ccKA and links it to phosphorylated DivK in the regulation of this process.
Of the ways in which pseudoenzymes regulate active enzymes, Elton Zeqiraj had a nice example : the inactive pseudoenzyme KIAA0157, which has structural similarity to deubiquitinating enzymes, and the ‘super-dimer’ it forms with the deubiquitinating enzyme BRCC45, thereby allosterically activating its cytoplasmic deubiquitinatase activity.
‘Zombie’ enzymes to inform studies of active enzymes, and how to find them!
Gerard Manning and Hesso Farhan disagreed on whether pseudoenzymes are best described as ‘dead’ proteins (Farhan) or ‘zombie’ proteins (Manning). However, both were clear that despite their varying degrees of vitality, their study has proved illuminating to the study of active enzymes .
Pseudoenzymes: best described as ‘dead’ proteins or ‘zombie’ proteins?
Manning’s talk on pseudokinases and pseudophosphatases, entitled ‘how not to phosphorylate a substrate and then not dephosphorylate it’, focused on using sequences and information on structural folds to predict activity of potential enzymes/pseudoenzymes, and using these structures to cast light on functions of pseudoenzymes shared with catalytically active enzymes. This came with a caveat that was later repeated by Nicholas Tonks: be wary of relying on sequence-based predictions alone, in that 14/24 pseudophosphatases were predicted to have no activity on this basis, but 4 have since been shown experimentally to have some.
Tonks’ talk was one of several on methods to identify which proteins are indeed pseudoenzymes. Rossanna Zaru spoke about the unique challenges of annotating pseudoenzymes correctly in the UniProtKB database to assist researchers, while Krysztof Pawlowski’s work on mining for new pseudoenzymes from sequences provided two new challenges: novel pseudoenzymes to study and a plea for the solution of more structures to help with this process!
Regulatory paralogs
A common feature of pseudoenzyme function that emerged from the meeting was the way in which inactive pseudoenzymes are capable of regulating active paralogs from which they were generated by gene duplication. Gustavo Afandor’s story on what he called ‘prozymes’ detailed the molecular mechanisms by which this occurs-in two paralogs of deoxyhypusine synthase, neither of which has significant activity on its own but only one of which is dead. Both are required for the regulation of polyamine synthesis in trypanosomatids.
This is particularly interesting since these organisms have no mechanisms for regulation of mRNA production, translation or stability, and therefore seem to have evolved a pseudoenzyme/prozyme-dependent regulatory mechanism instead!
Dame Janet Thornton spoke about the mechanisms of enzyme changes, gains and losses of function and the creation of such paralogs, as well as the use of the FunTree pipeline in categorising pseudoenzymes.
Natalie Jura spoke about the dimerization events involved in the complex allosteric regulation of the HER family which involve the catalytically impaired HER3 member, as well as tantalizing unpublished data which we cannot divulge!
Michael Ginger’s work on the mitochondrial histidine phosphatases, where a similar pseudoenzyme-enzyme interaction between two proteins was found by genome mining, raised interesting questions about the evolution of these enzymes and the functional relevance of their interaction: is it involved in mitochondrial signaling or a metabolic pathway?
Enzyme, pseudo-enzyme and ‘pseudo-pseudoenzyme’
Many speakers claimed to feel out of place, or even fraudulent, since their talks were on enzymes which actually had some enzymatic activity. However, all were great contributions. To coin a phrase, these ‘’pseudo-pseudoenzymes’’ fitted within the framework of the meeting: either because they were misclassified pseudoenzymes which retain functionally relevant, if low, enzymatic activity; or on account of having pseudoenzymatic functions and mechanisms despite being active enzymes.
Nicholas Tonks showed evidence that the predicted pseudophosphatase PTPN23, whose pseudoenzymatic functions in cancer had been discussed by Arnim Pause, was an active enzyme. This was a cautionary tale to all those tempted to make assumptions on function on the basis of sequence: typically inactivating mutations do not necessarily preclude function. Loss of PTPN23 was shown to promote tumour growth and invasion in breast cancer by increasing SRC phosphorylation and activation, showing at least some phosphatase activity was present in the enzyme.
PTPN23: a cautionary tale- typically inactivating mutations do not necessarily preclude function
A similar case was shown by Lincoln Potter in the case of the kinase homology domain of guanylyl cyclase enzymes. These proteins, important targets for blood pressure therapeutics, have complicated and only partially understood regulation, but by looking at the activation of these domains he showed how phosphorylated residues are required for this predicted inactive kinase domain to autophosphorylate. Their latest work showed further details of this elegant mechanism in which these phosphates create specifically charged patches on the kinase domain, which attach opposite sides of guanylyl cyclase homodimers to each other with salt bridges to regulate this signalling pathway.
The real frauds (as it were) of the meeting were distinguished speakers Susan Taylor and Sir Paul Nurse, with pivotal talks on PKA regulation and the molecular mechanisms involved and CDKs. Nothing pseudo about these enzymes- but the two talks were an arresting illustration of how studies of each inform the other to drive forward our understanding of complex cellular signalling networks.
- What is a Pseudoenzyme, and why? - 26th September 2016
- Great expectations: structural biology, macromolecular machines and moving targets - 27th May 2016
Comments