After watching CSI, and with forensic science being more advanced than ever, it’s easy to presume that criminals leave DNA traces everywhere that can help to make a conviction if they are caught.
Human hairs come to mind as a great place to start, however, the majority of samples recovered at crime scenes are shed hairs containing insufficient levels of nuclear DNA, meaning they cannot be used to make an identification. This is because short tandem repeat (STR) analysis is performed on crime-scene DNA, where probes are attached to the sample, then it is amplified in length by a polymerase chain reaction (PCR) to create a DNA fingerprint. Two samples can then be compared to find if there’s a match. With very little or no DNA present, this is not possible.
Furthermore, hairs aren’t actually left at crime scenes very often, with research suggesting that male pubic hairs are only transferred to the female genital area in around 4% of sexual assault cases. Finding alternative DNA-based methods would therefore be highly useful to increase the chance of identifying criminals.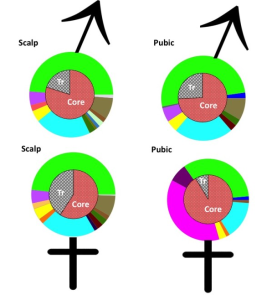 Research published today in the open access journal Investigative Genetics has proposed that the microbiome of bacteria hosted on our hairs could be used for human identification in forensic investigations. Specifically, they studied scalp and pubic hairs.
Research published today in the open access journal Investigative Genetics has proposed that the microbiome of bacteria hosted on our hairs could be used for human identification in forensic investigations. Specifically, they studied scalp and pubic hairs.
The researchers performed metagenomic analysis on 42 samples from seven individuals at three time points. They showed that each harbored unique microbial taxa that could be used to discriminate between individuals, and that pubic hair microbiota could determine between male and female samples based on the species present.
The only difference observed between male and female scalp hairs was the reduced level of transient bacteria on female samples, possibly due to increased ‘grooming activities’.
Interestingly, two of the study participants were a single co-habiting couple, and in one instance at the five-month time point the co-habiting couple’s microbiota were more similar to each other than previously. Interviewing revealed that the couple had sexual intercourse 18 hours prior to the collection of their pubic hairs.
This suggests that an exchange of bacteria had occurred, which the researchers say bodes well for future forensic applications involving sexual crimes. In addition, the authors concluded that bacterial colonies in pubic regions are more stable than scalp microbiota, making pubic hairs a preferred choice to consider further for forensic applications. This study is of course preliminary, and the authors propose that further investigation is warranted to look into the pubic hair microbiome as a forensic tool.
Previous microbiome studies have demonstrated that we all harbor unique bacterial compositions between body sites. It’s also been shown that our surface bacteria can be exchanged through bodily contact. An article recently published in Microbiome showed that a ten-second intimate kiss between individuals can shape the oral microbiota, involving the exchange of approximately 80 million bacteria. More regular kissing can lead to long-term colonization of microbes in the mouth.
 While advancing technology in forensic genetics hugely important for improving the conviction rates of criminals, the fact remains that a conviction cannot even be made purely on DNA evidence. Despite this, DNA fingerprinting still remains a hugely important tool in forensic genetics.
While advancing technology in forensic genetics hugely important for improving the conviction rates of criminals, the fact remains that a conviction cannot even be made purely on DNA evidence. Despite this, DNA fingerprinting still remains a hugely important tool in forensic genetics.
DNA fingerprinting was invented by Professor Sir Alex Jeffreys in 1984, through a “eureka” moment when he realised he’d found fully discriminative genetic markers in human blood. As part of a 2012-2013 thematic series on DNA fingerprinting Alec Jeffreys spoke with Investigative Genetics in a podcast about his invention of the technique, amongst other highlights during his scientific career, including the story of the first person to be exonerated of a crime based on DNA evidence.
There have been some past success stories from bringing microbial genetic analysis to the courtroom. A 2013 BMC Biology article describes a case where viral phylogenetics were used to prove the origin of a hepatitis C outbreak. Researchers modelled viral evolution rates to estimate the date of infection, tracking it to ten years before the outbreak and providing a connection between all of the cases.
An anesthetist was later convicted as being responsible for the infection of 275 of his patients with hepatitis C virus. In a BMC Biology commentary, Anne-Mieke Vandamme and Oliver Pybus comment on the importance of molecular phylogenetics as a tool for the forensic investigation of viral transmission.
Investigative Genetics is running an ongoing thematic series of research and reviews on microbial forensics. The series includes new guidelines by Bruce Budowle et al. for validation of high-throughput sequencing and microbial forensics applications. Sign up for alerts to receive updates when further articles are published!
Written by Sam Rose (@rosenovich), Journal Development Editor for Investigative Genetics
- Raising funds for genetic diseases - 23rd September 2016
- The Epigenetics and Chromatin Clinic - 9th November 2015
- Resurrecting one of the oldest genetics journals - 23rd October 2015
Comments