
Animal African Trypanosomiasis, also called Nagana, is a damaging and often fatal–unless treated–disease in animals. It is caused by several species of the Trypanosoma genus and according to the Food and Agricultural Organization of the United Nations (FAO), it is endemic in 37 sub-Saharan countries and kills an estimated cattle 3 millions each year, as well as affecting 100 million small ruminants. The economic and health impact it has in sub-Saharan Africa is huge, which is why the FAO has a special Programme Against African Trypanosomiasis.
A known challenge with trypanosomiasis diagnosis is that the parasite presence in the blood fluctuates with periods where it is detectable by microscopy and periods when it can not be detected using this technique. Current diagnostic tests vary in their sensitivity, specificity, ease of use and cost, limiting their use in screening and control. More research is needed to achieve a suitable diagnostic tool for veterinary use and One Health approach.
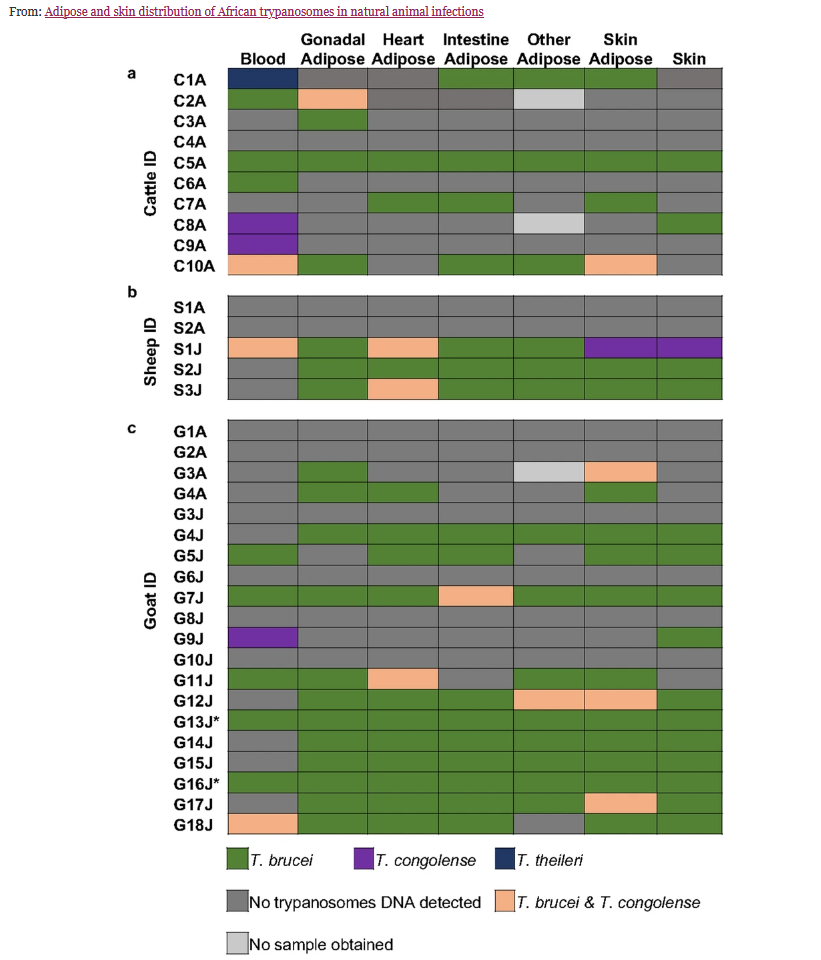
Although trypanosomes are widely thought of as blood parasites, they have been shown to invade different tissue types and, in experimentally infected animals, the parasite has been detected in skin and subcutaneous adipose tissues. In a recent study, researchers at the University of Ghana report on the distribution of different species of trypanosomes in naturally infected livestock, expanding the search to other adipose tissue samples.
The search for trypanosome species by tissue type in livestock
Researchers collected 242 samples from 35 animals, 20 goats, 10 cows and 5 sheep, in two government-approved abattoirs. The types of samples collected were:
- Peripheral blood from jugular veins
- Skin biopsies from neck region
- Adipose tissues from:
- the gonads
- heart
- intestine
- skin
- kidney
- and/or stomach
Genomic DNA was extracted from each of these samples and a nested polymerase chain reaction (PCR), which uses two sets of primers in two successive PCR runs to limit non-specific binding products, was used to identify positive samples and trypanosome species. The positive PCR samples were further characterized using Sanger sequencing. A Fisher’s exact test was used to compare the detected trypanosome infections between sample types.
It’s NOT always in the blood
Trypanosome DNA was found in 26/35 animals, and in 49.2% of samples (119/242). The most common trypanosome species was T. brucei, then T. congolense. There was one mismatch between the nested PCR approach and the sequencing data – the nested PCR had indicated one T. theireri infection however sequencing data identified this as T. congolense.
Regarding the types of samples, researchers found that there was no consistent pattern when testing for trypanosomes using different sample types – some animals had negative blood samples but positive for other tissues, whilst some were positive for trypanosome DNA in the blood but negative for other tissues, some were negative for both blood and tissues and others were positive for all. When they compared prevalence of trypanosome infections across tissue types they found that the highest number of positive samples were for skin adipose tissue and for gonadal adipose, whereas lowest was for blood. These were not found to be statistically significant.
Food for thought:
A key take away from this study is that parasitological and molecular diagnosis and screening of trypanosome infections in animals should use more than one sample type since trypanosomes DNA can be detected in skin and other adipose tissues even when it is absent from blood samples. This potentially can be a source of transmission to tsetse flies when then feed on animals and indicates that adipose tissues may be acting as a reservoir for these parasites in livestock and wild animals.
The authors of the study also suggest that the distribution of trypanosome species across samples may indicate a tissue preference by species, perhaps as a way of reducing parasite competition within the host.
The authors highlight that more research is needed on natural infections in animals to understand how host-parasite interactions may be influencing the distribution of trypanosome species within the host and for the development of effective screening diagnostic for African trypanosomiasis in animals.

Comments