
Pangolins are quite possibly the most adorable creatures on the planet. They are mammals (despite being covered in keratin scales), over eighty million years old as a species, and have inquisitive, intelligent personalities.

The pangolin has no teeth, and feeds off a diet of mostly insects using their long, thin tongues (longer than the length of their bodies) to probe ant and termite colonies. Pangolins have very few if any natural predators since their scales protect them by withstanding attacks from teeth and claws; a quick YouTube search will show lions trying and failing to eat a round shiny ball, who happily scampers away unscathed.
Unfortunately, the pangolins have been pushed to the brink of extinction by illegal businesses that market the meat and scales as “magical and medicinal”, and their rarity and cost has made them into a culinary status symbol in East Asian restaurants. This high market value has created a black market where pangolins have been cruelly poached from their natural environments of Africa and Asia to the point that all eight varieties of the pangolin are currently listed as critically endangered. Zoos have thus far been unsuccessful in keeping them, as very little is known about this shy and solitary animal.
Researchers Zelda du Toit and colleagues at the National Zoological Gardens of South Africa have set out to understand more of the mitogenomics of the four African varieties of the Pangolins; Temminck’s ground pangolin (Smutsia temminckii), the giant ground pangolin (S. gigantea), the white-bellied pangolin (Phataginus tricuspis) and black-bellied pangolin (P. tetradactyla).
Once the full mitochondrial DNA genomes from these specifies and subspecies has been sequenced and verified, researchers will hopefully be able to assist in providing a better understanding of the evolution of pangolins, which in turn will be essential for conservation genetic studies.

Mitochondrial DNA is well suited for evolutionary studies due to its high mutation rate, well-structured genome with restricted non-coding DNA sequences, and lack of recombination.
Du Toit and colleagues set out to perform next generation sequencing for all four African pangolins using Illumina HiSeq 2500 in order to reconstruct complete mitochondrial genomes, two of which have previously not been sequenced; the black-belied pangolin and the giant ground pangolin. Du Toit and her team of researchers have also managed to conduct a phylogenetic assessment in order to provide a genus level classification of African pangolins.
By using six deceased pangolins which comprised of the four African subspecies, full mitogenomes were generated and aligned with eleven other carnivore genomes using MAFFT v7, as they are reported to be evolutionary closely related to pangolins. The best fit model of sequence evolution for the dataset under the AIC was the General Time Reversal model (GTR+I+G) with invariable site and gamma distribution of 0.822, and the best fit model under the BIC and DT was the Transition model two (TIM2+I+G) with invariable site and gamma distribution values of 0.998 and 0.990, respectively.
From these two models, three Bayesian phylogenetic trees along with three maximum likelihood trees were generated for the datasets. The different trees all showed consistent branching patterns, posterior probability values for the BI trees and bootstrap values for the ML trees.
From the three trees, a consensus tree was created, as seen in Figure 2 of the study:
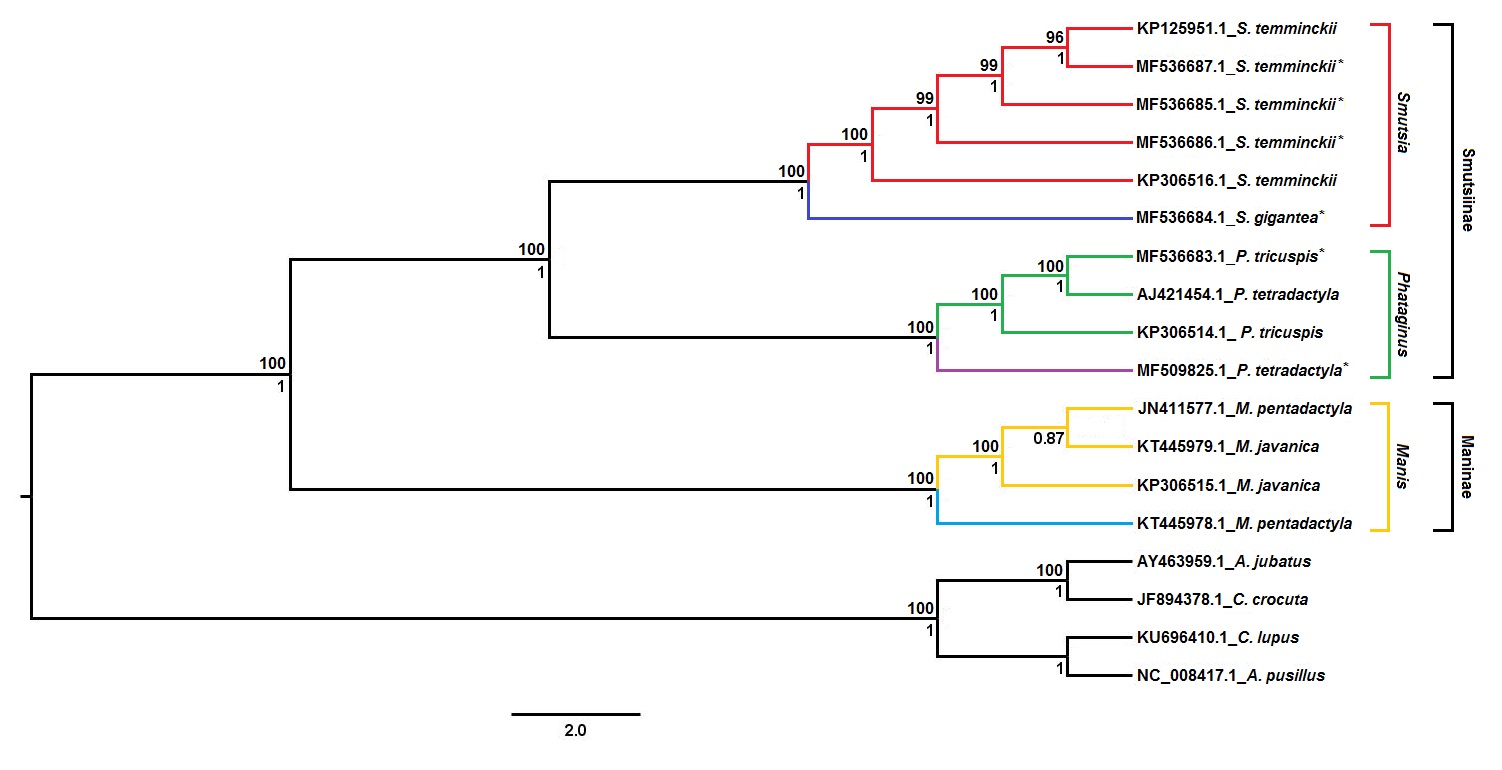
From this tree, it can be deduced that all African pangolins group according to the species, with the exception of the black-bellied pangolin genome. That mitogenome has previously been reported to be misclassified based on partial Cob analysis and was confirmed in the analysis as a white-bellied pangolin genome. Additionally, the phylogenetic tree in figure 2 provides support for the separation of Asian and African pangolin species into two distinct monophyletic clades with the latter consisting of all four African pangolins species. Within the clade, the giant and Temminck’s pangolin cluster separately from the white-bellied and black-bellied pangolins with significant Bayesian and ML support.
The authors further go on to claim that the observed branching pattern provides support for the classification of the group-dwelling and arboreal species into two separate genera; Phataginus and Smutsia. Results from their analysis also suggest the overall classification of pangolin should be in three genera: Manis (Asian pangolins), Phataginus (African tree pangolins), and Smutsia (African ground pangolins). However, the authors caution that further analysis of the Asian pangolin genome is necessary. The branching patterns were also confirmed using individual loci.
This study is very unique in that it sheds a light on the diversity of the endangered pangolins remaining in the world. By discovering two new reference genomes for the black bellied pangolin and the giant ground pangolin, researchers will be able to better understand the evolutionary diversity of the extant pangolin species. Additionally, this study offers the first phylogenetic assessment of six of eight species using whole mitochondrial DNA genomes, and the data suggests that the African and Asian pangolin species are indeed separate monophyletic clades, and there is further separation of genera between tree-dwelling and ground-dwelling African pangolins.
These studies are essential for the conservation efforts of this species, and hopefully will be able to help boost dwindling wild pangolin numbers before they go extinct; this species has existed for over 80 million years, and to lose them now would be to lose a very unique and majestic animal.

3 Comments