DNA barcoding is now a standard part of the biodiversity toolkit. Nearly 20 years ago, the idea arose – simultaneously in several groups – that advances in DNA sequencing might make it possible to identify species cheaply and efficiently using small parts of their genomes. Despite early scepticisms, the DNA barcoding movement, driven by Paul Hebert and colleagues at the Biodiversity Institute of Ontario, has been a runaway success. It is now routine to generate a standard DNA barcode of ~650 DNA letters for every specimen. Over 6 million DNA barcodes from over 260,000 species have been logged in the Barcode of Life database (https://www.boldsystems.org/).
Having all the data in the central Barcode of Life database is a revelation: while 260,000 recognised, named species have DNA barcodes attached to them, there are nearly 500,000 different clusters (sets of sequences similar to each other but distinct from anything else) of DNA barcodes in the database. The quarter-million extra clusters are likely to be real, new species – organisms that have not yet been formally named. The huge number of undescribed species is not a huge surprise to biodiversity scientists, but is challenging to biodiversity science.
Three things about these new putative species stand out. One is that many are “cryptic”. This means that they were collected and initially identified as being members of a known species, but were found to be distinct based on their DNA barcode. So they must be very similar in morphology to known species. A second is that many are very small – tiny insects and the like. It looks like the biodiversity we have missed is in the vast majority of life that is best seen under a lens. Lastly, many of these species were found in areas where no new species are expected – in the back gardens (literally) of respected biodiversity scientists, in fields and parks where naturalists have been collecting for centuries. Many do come from undersampled tropical forests, but equally fruitful are urban shrubberies. New species are everywhere.
We work on nematodes, or roundworms. Most nematodes are tiny, live hidden in the soil and marine muds, and are difficult to identify. They are also extraordinarily abundant. You definitely need a good guide, a good microscope, and a lot of patience. Marie-Anne Félix, at the Institute of Biology of the Ecole Normale Supérieure in Paris, started a tiny revolution a decade ago when she went looking for relatives of the most famous of nematodes, the Nobel prize-winning Caenorhabditis elegans. (Strictly, the Nobel prizes were given to the humans who made huge discoveries using C. elegans, but “the worm” was up there on the podium with them…)
Many of the nematodes she encountered were new species, but morphologically cryptic. She could tell them apart because they would not interbreed with any of the other Caenorhabditis species she kept in culture. Through the efforts of Marie-Anne and the other collectors she inspired, there are now over 50 species of Caenorhabditis in culture. Most of these species have not been formally named, but all are tiny worms (~1 mm long), see-through, and hard for humans to distinguish – except by DNA barcoding.
Formally naming these species is important, as it registers them in the matrix of scientific knowledge, and provides a hook on which to hang discoveries and comparisons. So the community of Caenorhabditis diversity enthusiasts is now doing just that. Karin Kionkte (New York University), Marie-Anne and colleagues published a radical paper in 2011, describing 15 new species, including a diagnostic DNA barcode marker for each [1]. This paper included minimal morphological descriptions – simply affirming that the species were Caenorhabditis and describing the male tail.

A single DNA barcode marker may not always be enough to define taxa, and is by definition from a gene that is found in all species and so unlikely to tell us anything about novel biology. Much more useful would be a catalogue of “all” the genes of a species, conserved ones as well as new ones, in the context of the whole genome… In the decade or so since the first major publications on DNA barcoding, the technology of DNA sequencing has been transformed. It is cheaper and faster than ever before. So we decided to sequence the whole genomes – all 100 million DNA letters – of all the Caenorhabditis species we could. We called this idea the Caenorhabditis Genomes Project (CGP), invited Marie-Anne and others to join, and started sequencing…
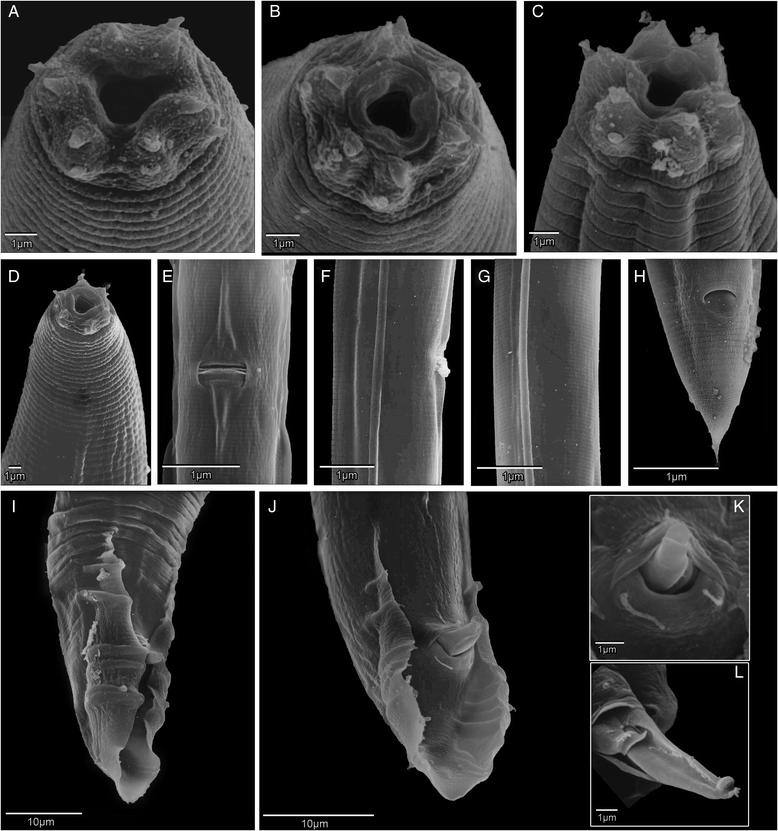
One of these Caenorhabditis species – informally called “C. sp. 1” – was originally discovered living in the bracket fungus Ganoderma applanatum growing on a stump of wood by a student of Walter Sudhaus (Freie Universität Berlin). From 2013 onwards, Dieter began exploring the woodlands of Europe, eventually isolating several more strains of C. sp. 1, mostly from the same species of fungus. He then hooked up Walter Sudhaus in Germany and agreed to collaborate to describe the species. Karin Kiontke kindly supplied a sample of the original strain to Dieter for comparison, who then worked up a formal, morphological and ecological description of C. sp. 1, and we collaborated to provide DNA barcode data – the whole of the genome. Our paper “Caenorhabditis monodelphis sp. n.: defining the stem morphology and genomics of the genus Caenorhabditis” was published in BMC Zoology and formally describes C. sp.1 along with its entire genome sequence.

Microbiologists are now used to publishing full genome sequences along with descriptions of new bacterial species – since most bacteria have small genomes (less than 7 million DNA letters) this is quite easy to do nowadays. The tide is changing, though. A fungal species has been formally described with its genome sequence. And the genome of a strepsipteran insect (a wonderfully weird group of parasites) was published separately but simultaneously with its species description. We think C. monodelphis is the first time the whole genome of an animal has been a part of the species description. We think this is the “way to go” for the future: describe a species – describe its genome.
Why study C. monodelphis? C. monodelphis is a key species for understanding the evolution of all Caenorhabditis. When we placed it on an evolutionary tree – using whole genome data from the many species we have sequenced as part of the CGP – we found it was sister to all other known Caenorhabditis species. This means we can use it to estimate what the genome, and thus the biology, of the ancestor of all Caenorhabditis might have looked like – and answer (some of) the question of how the model species C. elegans came to be just as it is.
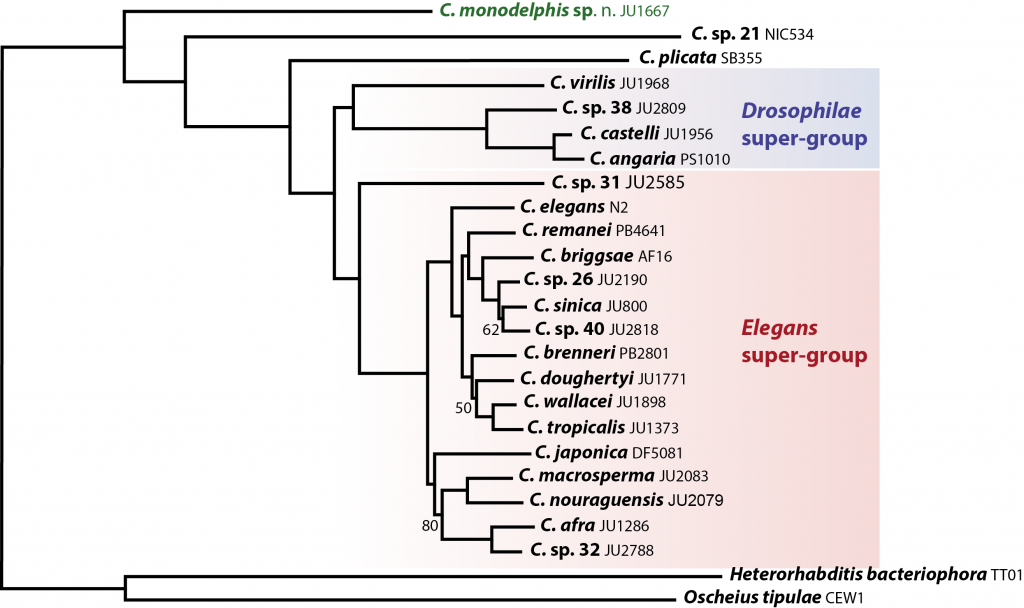
Our analyses of the genome are still in their early stages, but we have already found some striking differences compared to C. elegans. For example, genes in C. monodelphis have nearly twice as many introns as do the same genes in C. elegans. Given our other data, we think this means that the Caenorhabditis ancestor was intron-rich and that there has been extensive loss during the evolution of C. elegans. But we don’t know why C. elegans has lost introns – yet. Analysis of all the other genomes in Caenorhabditis will allow us to test ideas of what drivers and mechanisms are at work, and these drivers and mechanisms may turn out to be true of many other species too. This is precisely what we hope the CGP will provide: an essential evolutionary context for C. elegans and the wealth of research associated with it.
The Blaxter lab already have another six Caenorhabditis species to publish with their genomes as the ultimate DNA barcode, and we will soon be half-way to our goal of sequencing all Caenorhabditis – as long as Marie-Anne, Dieter and their colleagues don’t keep on discovering new species. Doubtless they will, and that will keep us (happily) busy [3].
———————————————————————-
Thanks to the Blaxter lab and colleagues in the CGP for support. We exploited the twitter following of Sujai Kumar (@sujaik) to find out who else has published a genome alongside a eukaryotic species description.
Genomic data for C. monodelphis (and for all the other Caenorhabditis species we are sequencing) are available to browse, query and download at https://www.caenorhabditis.org.
A species description of a ergot fungus alongside its genome
Chen L, Li XZ, Li CJ, Swoboda GA, Young CA, Sugawara K et al. Two distinct Epichloë species symbiotic with Achnatherum inebrians, drunken horse grass. Mycologia 2015:863-873.
A species description of the strepsipteran Mengenilla moldrzyki and its genome published in separate publications, but concurrently
Niehuis O, Hartig G, Grath S, Pohl H, Lehmann J, Tafer H et al. Genomic and Morphological Evidence Converge to Resolve the Enigma of Strepsiptera. Curr Biol 2012;22:1309-1313.
Pohl H, Niehuis O, Gloyna K, Misof B, Beutel R. A new species of Mengenilla (Insecta, Strepsiptera) from Tunisia. ZooKeys 2012;198:79-102.
Transcriptome data published alongside a species description
Edmunds SC, Hunter CI, Smith V, Stoev P, Penev L. Biodiversity research in the “big data” era: GigaScience and Pensoft work together to publish the most data-rich species description. GigaScience 2013;2:14.
References:
[1] Kiontke KC, Felix M, Ailion M, Rockman MV, Braendle C, Penigault J & Fitch DHA. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evolutionary Biology 2011; 11:339.
[2] Slos D, Sudhaus W, Stevens L, Bert W and Blaxter ML. Caenorhabditis monodelphis sp. n.: defining the stem morphology and genomics of the genus Caenorhabditis. BMC Zoology 2017; 2:4.
[3] Blaxter ML. Imagining Sisyphus happy: DNA barcoding and the unnamed majority. Philos Trans R Soc Lond B Biol Sci 2016; 5:371(1702).
Comments