
Plasmodium falciparum is familiar to many as the parasite that causes the most severe form of malaria. Malaria is responsible for about 200 million infections and at least half a million deaths every year, most of the latter in children under the age of 5. One of the reasons why infections are so serious is the ability of the parasite to sequester in blood vessels and also evade the immune response of its host, continuing to multiply within a hostile internal environment.
A key survival factor for Plasmodium in its human host is the protein PfEMP1 (Plasmodium falciparum erythrocyte membrane protein 1). This is exported to the surface of infected red blood cells, causing them to adhere to the endothelial lining of blood vessels and preventing their transport to the spleen and subsequent destruction (along with the hitch-hiking parasite). Unfortunately, sequestered erythrocytes (particularly in the brain) can obstruct blood vessels, resulting in complications such as haemorrhage and hypoxia, and leading to the most severe manifestation of the disease, cerebral malaria.
The importance of PfEMP1 to parasite survival makes it a crucial target for the immune system;
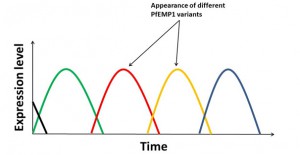
however, Plasmodium has evolved a way to evade the immune response by producing and switching between different PfEMP1 variants. Thus when one variant is targeted by the immune system a different variant – not yet recognised – becomes the dominant expressed form and allows the infection to persist.
PfEMP1 proteins are encoded by a large multigene family called var comprising ~60 different genes each producing an alternative PfEMP1. Recombination processes in these genes also allow for the creation of new variants, granting the parasite many millions of potential PfEMP1s.
The relationship between var expression and antigenic variation of PfEMP1 is well-established but the processes which govern switching and ensure expression of only a single variant at any one time are less well understood. In general it is thought that var gene expression is governed by epigenetic mechanisms; that is to say by factors marking var-encoding chromatin and leading to genes being heritably switched on or off. For example, the acetylation and methylation state of histones can dictate whether individual var genes are ‘off’ or ‘on’ and there may also be a dedicated ‘permissive site’ within the nucleus which only a single var gene may occupy at any one time. A full commentary on this area of research in Plasmodium can be found here.
A recent paper by Zhang et al contributes a further possibility by describing a previously unknown process of expression control via degradation of var gene RNA transcripts. Zhang and colleagues used bioinformatic methods to identify a novel exoribonuclease called PfRNase II. This protein appeared not to belong to the complex that controls standard RNA processing, maintenance and degradation (called the exosome) but it did degrade single-stranded RNA, validating it as an RNase.
The researchers were unable to knock out PfRNase II without affecting parasite survival, and inferred that the gene therefore had an essential function. Instead, they employed an alternative method that allowed them to temporarily degrade the protein without inactivating the endogenous gene. By fusing a destabilization domain (FK506-binding protein, FKBP) to the end of PfRNase II, the researchers could control whether or not the protein was degraded by adding or withholding a protective ligand. In fact, the tagged protein maintained only some of its usual functions, allowing these parasites to survive, but without their normal control over var gene expression patterns (the addition of a large tag can often in itself disrupt an enzyme’s normal function).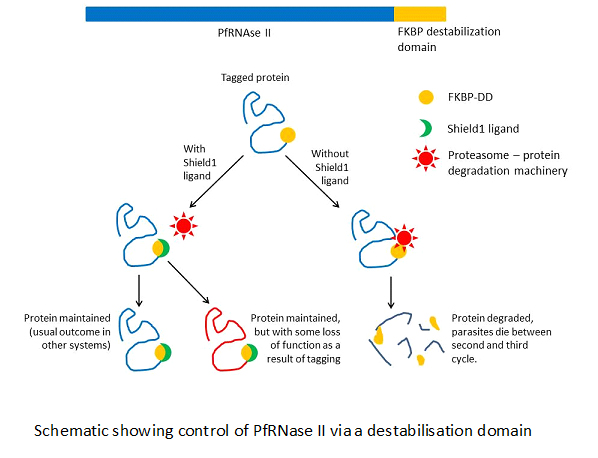
The researchers then used this system to investigate patterns of transcription with active or inactive PfRNase II. RNase-inactivated parasites substantially upregulated 213 genes, almost all of which were from variant virulence gene families (e.g. var) or from adjacent genomic regions. There was also a particular connection with a subset of var genes known as upsA which were very highly upregulated, despite being rarely expressed in vitro.
upsA vars are of particular interest because their expression is associated with the most severe manifestations of malaria in the field. Indeed, Zhang et al. analysed transcript levels from patients and found, as previously reported, that levels of upsA transcripts were higher in patients with severe as opposed to uncomplicated disease. Importantly, this correlated with low levels of PfRNase II expression, suggesting that low levels of RNase activity may directly promote the switching on of severe-disease-associated antigens.
Most significantly of all, parasites without PfRNase II activity lost the ability to express only a single var gene at a time, with several variants from both upsA and upsC subgroups co-expressed. This suggests that PfRNase II is responsible for silencing var genes by constantly degrading RNA transcripts and, by extension, that var genes are constantly producing short-lived transcripts – a state which may prime the parasites for rapid antigenic switching. This fits with an earlier observation that upsA var genes are the most ‘labile’ of the var family – switching on readily in patients but also switching off readily in vitro. A key question for the future will be what controls the controller: does the level of PfRNase II vary at random during human infections, or might it be responsive to the human host environment?
Finally, this mechanism may also apply to other parasites that singly express one of an array of variant antigens, a common feature of many protozoan parasites including Giardia and Trypanosoma; or indeed any other organism which employs single expression from a panel of multiple variants. It will be fascinating to discover how widespread and conserved such a mechanism might be. As to prospects for improved treatment of malaria, PfRNase II may prove to be a valuable tool to manipulate the expression of parasite virulence genes or indeed – since it is essential for parasite survival – an important new target for therapy in its own right.

Comments