
A rare disease, also referred to as an orphan disease, is any disease which affects a small percentage of the population, with the majority of such diseases being caused by altered functions of single genes. Although individual diseases may be rare, collectively they are common, affecting millions of people worldwide. The Global Genes Project estimates that a staggering 300 million people worldwide are affected by a rare disease.
However, whilst there is a wealth of research surrounding rare diseases in humans, in livestock animals rare diseases are traditionally poorly diagnosed. In non-laboratory animals, the number of rare genetic diseases is unknown; however, Online Mendelian Inheritance in Animals (OMIA), a catalog of inherited disorders and associated genes in animals, reports more than 2500 phenotypes in eleven domestic animal species.
Other than clinical description and pathological examination, the underlying genetic causes have, for the most part (approx. 80%), remained unknown. Either recessive inherited mutations which occurred generations ago or spontaneous de novo mutations can be responsible.
Thankfully, substantial progress in the field of veterinary genetics has been made In the last decade. Many domesticated animal genomes, including the bovine genome in 2009, have been sequenced. Recently, the 1000 bull genomes project has been initiated and allows participants worldwide to access sequence data from a large number of cattle control genomes. With sequencing costs also decreasing rapidly, the combined usage of these new resources and tools has revolutionized the dissection of the genetic basis of diseases and traits that have proven to be intractable to conventional gene-discovery strategies.
 Contributing to the growing knowledge of veterinary rare diseases, new research published in BMC Veterinary Research presents the findings from a successful collaboration between both genetic and pathology experts in Germany and Switzerland.
Contributing to the growing knowledge of veterinary rare diseases, new research published in BMC Veterinary Research presents the findings from a successful collaboration between both genetic and pathology experts in Germany and Switzerland.
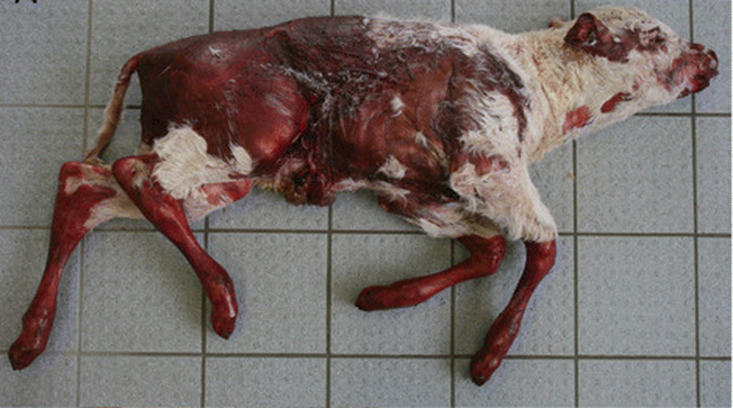
Using state-of-the-art methods, the team of investigators were able to determine the genetic background of a rare skin disorder, epidermolysis bullosa junctionalis (EBJ), in a Charolais calf – an important beef breed originating in France.
Epidermolysis bullosa is the umbrella term describing a group of congenital and hereditary connective tissue diseases that result from defects in the dermal-epidermal attachment structures. There are three main types of epidermolysis bullosa, including EBJ, but each cause the skin to become very fragile, resulting in painful blisters in the skin and mucosal membranes. In humans, several causal mutations for EBJ have already been identified; however, the causal mutation for EBJ in Charolais cattle remained unknown.
In this study, the authors hypothesized the defect in the calf to be either inherited in a recessive fashion, or to be caused by a dominantly acting de novo mutation.
As there are many diseases and congenital syndromes known in domestic animals, where one cannot find similar diseases in other species, the whole genome sequencing approach is currently used to detect spontaneous de novo mutations in single affected animals.
Therefore, to identify the causal mutation, the authors used next-generation sequencing technology to sequence the whole genome of the affected animal. Here, the sequencing data is checked for deviations from the reference sequence, giving some thousands of sequence variants across the whole exome, the subset of the genome that is protein coding, in one animal.
In this current study, a search for such variants in the recent sequence variant database of the 1000 bull genome project database containing already sequenced ‘normal’ cattle genomes was conducted. This ‘discrete-filtering’ step is used to eliminate mutations in genes by assuming that any variant found in the ‘filter set’ cannot be causative. Thereby, the number of unique (private) DNA variants present in only one affected animal could be reduced to a very small number of high-priority candidates (if not a single one).
However, although the use of advanced technology was fundamental to the study, it is of importance to note that a detailed characterization of the disease phenotype was an indispensable prerequisite for the authors.
Looking for similar defects in human medicine finally helped the team to identify the causative mutation for this fatal bovine skin disorder. After screening for coding variants, they identified a concordant loss-of-function mutation in integrin beta 4 (ITGB4). This is the first time in which a recessively inherited ITGB4 associated EBJ has been reported in cattle.
Overall, this study serves as a reminder that current DNA sequencing methods now offer a powerful tool for understanding the genetic background of rare diseases in domestic animals. For breeders, being able to identify causative mutations in rare diseases of livestock allows us to select against these known mutations, helping to eliminate such disorders in animal production systems.

Comments