Having been quite versed in the art of research 8 years post PhD, I have been very fortunate to witness a renaissance in publishing in two ways. First, I remember quite well during my PhD training (over 10 years ago), the process of preparing a manuscript for the highest ranked journal—submit, reject, reformat and submit to the next journal, reject, submit . . . you get the story. During that time, Impact Factor was the key metric in which a journal was measured. This evolved quickly into a measure of a researcher’s performance. There was active push to get manuscripts into journals with a high Impact Factor.
Over the course of my career, merit in using the Impact Factor to judge a study was questioned. These days, it is almost a profanity to even consider the Impact Factor as a measure of a manuscript’s value (Randy Schekman’s Piece on Nature, Cell and Science). Believe it or not, I have heard a swear jar was enforced at a grant review panel, where whomever mentioned “Impact Factor” would contribute to the swear jar.
Back to my inaugural first-author manuscript, I aimed my submissions to the major journals including Science, Nature, Cell, Nature Structural Biology, Developmental Cell, each time sacrificing some Impact Factor points. This was at my PhD supervisor’s guidance, as we both thought of our body of research to be paradigm shifting at the time. I quickly won the boomerang award for the manuscript that came back the most times within the group (not exactly the prize worth shouting about!). In the end, we settled on, what was at the time, a very new publishing house, the Public Library of Science (PLoS). “What is this open access business? No business if you ask me; how will they survive if you give everyone open access to their published papers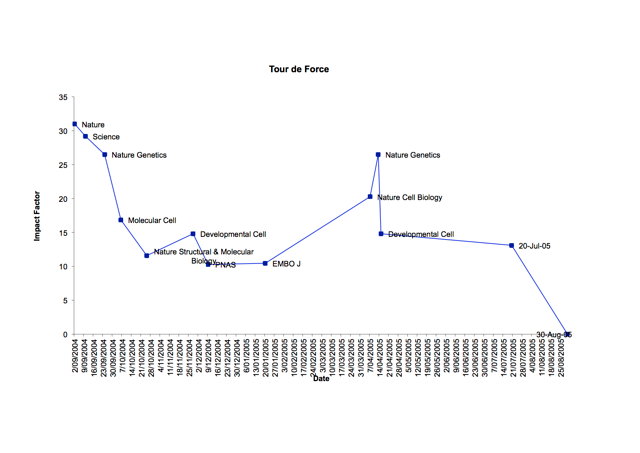 ?”
?”
But I was sold on open access. It took a little convincing for my supervisor to come around, who settled on the analogy with the stock market, “well the Impact Factor can only go up with this one”. To make a long story short, my paper was published in PLoS Genetics. It was an interesting finding to me and my supervisor, but wasn’t really to the editors of Nature and Science. Today, open access publishing has grown, with the first open access publisher BioMed Central (BMC) now publishing over 260 journals spanning all topics of biology and medicine. Publishers like BMC and PLoS paved the way for some of the newer publishers like PeerJ and eLife.
As scientists, we all have come to realise the benefits of open access. Seeing the power to quickly disseminate research papers to the research community much quicker than conventional print-based avenues, Nature and Science have also embraced open access. Measures of impact have evolved in the era of open access; however, open access is only slowly being adopted by the important funding bodies. It has become a requirement by NIH in USA, and more recently in Australia NHMRC and ARC and the Chinese Academy of Sciences, that government funded research should be published as open access. Moreover, philanthropic bodies are also starting to require research be published in open access journals.
Reproducible Research
Reproducible research is the essence of scientific endeavour. I feel the greatest form of flattery to be had of a scientist is a peer whom replicates their findings. I came across the concept at a CSIRO Computer and Simulation Sciences eResearch Conference (CSS) held in Melbourne earlier this year. The organisers invited Victoria Stodden and her talk on reproducible research was the one talk I took away from the conference as the issue of the moment, particularly in omics and big data analytics.
There are many examples of genomics analysis whereby the details on the analysis of data are not fully articulated in the published manuscript. This is a huge problem with regard to reproducible research because without the methods, it is almost impossible to replicate the results of the study. Publishers are starting to address this issue. GigaScience is at the forefront and one of the pioneers in publishing data, assigning DOIs to datasets and the methods which the data was derived by. Moreover, there are many online tools to facilitate the recording of software, algorithms and scripts used to process “omics” data. These include iPython Notebook, Galaxy and knitr.
Doin g something about it – The G3 Workshop
g something about it – The G3 Workshop
The more I delve into this important area of open access, open data and publishing, the more I find that the stakeholders and the scientists that need to publish their work should be well aware of and exploit to their full potential open science initiatives and tools. In light of this, I have been involved with putting together a workshop in Melbourne, Australia, called the Great Galaxy GigaScience (G3) Workshop.
This one-day event will be held on September 19 at The University of Melbourne. Two concurrent workshops will take place, the first on Galaxy, a web-based platform for bioinformatics analysis while the second is on Authorea, an online manuscript drafting facility that is great for collaborative writing (with Git running under the hood for version control). This along with talks from some leading scientists on the top ten rules for presenting data by Professor David Vaux, as well as members of the GigaScience Editorial and Data Team.
In formulating this wor kshop I have co me together and worked with some great people including those at Melbourne ITS Research and VLSCI. I have also come across some cool new tools that will certainly help my publishing prowess and acumen. The G3 workshop would not have come about without the support and input from GigaScience and financial sponsors including the Australian Bioinformatics Network, Victorian Life Sciences Computational Initiative (VLSCI), Illumina, Research Bazaar and The University of Melbourne.
kshop I have co me together and worked with some great people including those at Melbourne ITS Research and VLSCI. I have also come across some cool new tools that will certainly help my publishing prowess and acumen. The G3 workshop would not have come about without the support and input from GigaScience and financial sponsors including the Australian Bioinformatics Network, Victorian Life Sciences Computational Initiative (VLSCI), Illumina, Research Bazaar and The University of Melbourne. 
One Comment