Drug response and risk of adverse reactions are influenced by genetically inherited and acquired inter-individual variability in drug absorption, distribution, metabolism and excretion. This causes large differences in how different subjects respond to drugs and the extent to which they suffer from side effects.
Until now, pharmacogenomic biomarkers for common genetic variants have been used as tools in drug development and in clinics for patient stratification and prediction of the specific drug metabolism and response exerted by individual patients. However, the genetically encoded variability is much higher than can be predicted for by these biomarkers.
This is explicitly illustrated in twin studies that demonstrate that only 40–45% of the inherited variability in the pharmacokinetics of two different commonly prescribed drugs can be explained by the known common mutations. Research by us and others has revealed that patient specific rare mutations are important contributors to this unexplained heritability and might explain 20–40% of the overall inter-individual variability in drug response.
Thus, there is a need for tools that utilize the genetic information of a patient more comprehensively in order to further personalize drug treatment. Such an approach should principally consider all mutations affecting metabolism and response to a given drug in a specific patient, which requires information about the patient’s genomic makeup.
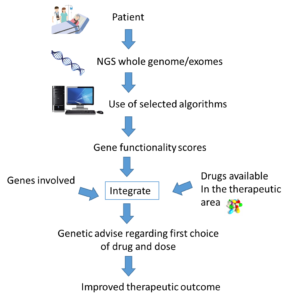
While the development of new sequencing protocols and the rapid decrease of sequencing costs drastically reduce the technical hurdles of such an approach, the major problem is how this information should be translated into specific clinical advice. How do we know which among the hundreds of identified mutations cause functional variation and are important for specific drug treatment?
In an article recently published in Human Genomics, we analyzed sequence differences in 208 different genes having influence on variability in drug metabolism, transport and response (pharmacogenes). By analyzing exome sequencing data from more than 60,000 individuals, we found that the pharmacogenetic architecture was highly complex with each individual harboring an average of 41 functional variants; rare variants accounted for 11% of these.
The relative importance of rare variants was highly gene and drug-specific. While the functional variability of some genes was governed by few high-frequency variants, rare variants accounted for the entire genetically encoded variability for more than half of the pharmacogenes. The analyses of mutations in regulatory regions was lacking here, which is estimated to account for about 1–2% of the number of variant alleles based on the CYP genes.
The functional impact of the mutations was analyzed using a computational prediction framework specifically developed for pharmacogenes through analysis of high-quality experimental data for pharmacogenetic variants with >90% sensitivity and specificity.
We indicate using five different commonly prescribed drugs that rare exonic variants account for a substantial part of the unexplained inter-individual differences in their metabolism. Importantly, these analyses suggest that Next Generation Sequencing-based approaches coupled with computational variant interpretations can refine personalized drug response predictions and incentivize the testing of this concept in clinical trials.
In conclusion it appears evident that we can get predictions of the functionality of pharmacogenetic mutations with reasonable accuracy, allowing for personalized activity scores for all genes with relevance for a given drug treatment to be derived. Instead of single SNP variations, all functional mutations in each gene should be summarized into a gene functionality score. Importantly, based on these gene functionality scores in combination with other patient-specific factors, we can formulate the most beneficial drug treatment with respect to choice of drug and dose.
In the future we anticipate that sequence data will be available preemptively, facilitating its clinical implementation. This would lead to safer, more efficient pharmacological therapy, improved public health, and reduced societal costs for drug treatment.
Comments