
The situation
Schizophrenia is a chronic mental disorder that manifests in adolescence and often leads to a lifetime of impairment and permanent disability. Schizophrenia is characterized by widely divergent symptoms, from hallucinations to lack of motivation to lack of attention. These different courses of disease progression suggest heterogeneity of underlying biological causes of schizophrenia.
The problem
Despite decades of scientific efforts, schizophrenia lacks an objective biological diagnostic test. Instead, the diagnosis of schizophrenia still exclusively relies on the subjective clinical interview between patient and clinical psychiatrist.

This creates a problem–how can the underlying biology of schizophrenia be investigated when the investigation begins with an inherently non-biological concept: a subjective clinical diagnosis?
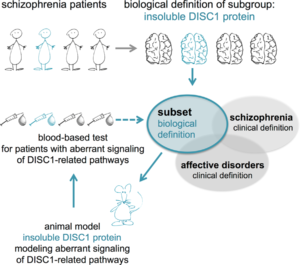
The problem is further exacerbated by the method of classification. There are no clinically approved subsets of schizophrenia. Therefore, even if significant biological findings in a small subset of schizophrenia are discovered, they will be diluted, because they are pooled with all clinical cases of the disease.
However, if starting with the clinical diagnosis is too broad and not biologically accurate, how do we go about detecting subsets of schizophrenia?
The solution
In our recent study in Translational Psychiatry, we worked to resolve the Catch-22 created by the interaction of the subjective and objective. We began by designing an animal model that would exhibit symptoms akin to those seen in schizophrenia patients, namely the disruption of certain cellular signaling pathways in the brain.
We decided to focus on a protein called ‘Disrupted-in-schizophrenia 1’ (DISC1). DISC1 is a hub protein critical to neuronal development and the maintenance of adult nerve connections (synapses). We genetically engineered a rat model where the DISC1 protein was dysfunctional.
The transgenic rat displayed abnormal behavior, incorrect neuronal connections, and an imbalance of neurotransmitters–all symptoms reminiscent of behavioral disorders like schizophrenia.
Armed with an animal model with the same aberrant signaling networks as schizophrenia patients, we identified unique blood markers in the model, as these same blood markers may identify where similar molecular pathways might be disturbed in schizophrenia patients. From there, we would be able to stratify patients independent of clinical diagnosis.
We identified such blood markers in the rat model by investigating differential gene expression blood cells and discovered that they belonged to a larger, dysregulated network of immune-related genes. When we tested the top markers, termed RGS1 and CCL4, in two independent cohorts of schizophrenia patients, they were equally diminished in both cohorts.
With the help of these two markers representative of a dysregulated immune network, we could finally identify a subset of 25% of clinically defined schizophrenia patients with 97% specificity. In other words, by dysregulating this immune-related network of genes, we were able to definitively define a subset of schizophrenia.
Next steps
Our study presents a unique pairing of an animal model for a subset of schizophrenia and a blood test for identifying the corresponding patient subset. The schizophrenia subset we identified might display similar biological irregularities as the corresponding animal model. If these irregularities correspond, the study of the animal model may allow for the development of causal pharmacotherapies effective in this specific schizophrenia patient subset. Our study illustrates a novel and innovative way to identify subsets of mental illness patients by reverse translation, potentially opening the door to a huge change in the diagnostic culture in clinical psychiatry.
Enjoyed this blog post? Read the full article in Translational Psychiatry.
Comments