At the beginning of September 2013 I weighed about 205lbs (92kg). I decided to do something about my weight, for my health and for the sake of my family and of course I approached this plan as a scientist. For me that meant parameterizing my inputs and outputs so I could control what I was doing to my body.
I bought a FitBit, a pedometer that calculates many aspects of my activity and calorie burn, and also started to monitor the calorific content of my food intake.
I had several plans that involved distributing calorie intake differently across the day, including more calories at breakfast, and less at lunch and dinner; however, primarily, I made sure I was always burning more calories than I was eating. Within 12 weeks I has lost 40lbs, just in time for Thanksgiving!
As a microbial ecologist, I know that aspects of this bodily change were significantly influenced or were influencing the microbial organisms that call me home.
Indeed, while I was not diligent with my microbial profiling, the bacterial community in my gut did change. But as I changed so many things about my life, so rapidly, it is impossible to determine which of these changes influenced my shifting microbial gut the most. Also, I am only one person! A statistical analysis cannot prevail with only a single individual; we need more participants to determine if an observed correlation is meaningful.
The issue of inter-personal comparison is compounded by the individuality of the microbiome. This individuality fascinates me. The fact that my microbiome differs significantly from your microbiome, and that I can use that information to differentiate and identify you from me, is both interesting and has some incredible potential.
Pioneering work by Eric Alm and colleagues at MIT, published today in Genome Biology, provides evidence that supports the individuality of the microbial signal, and more importantly points to its stability over time.
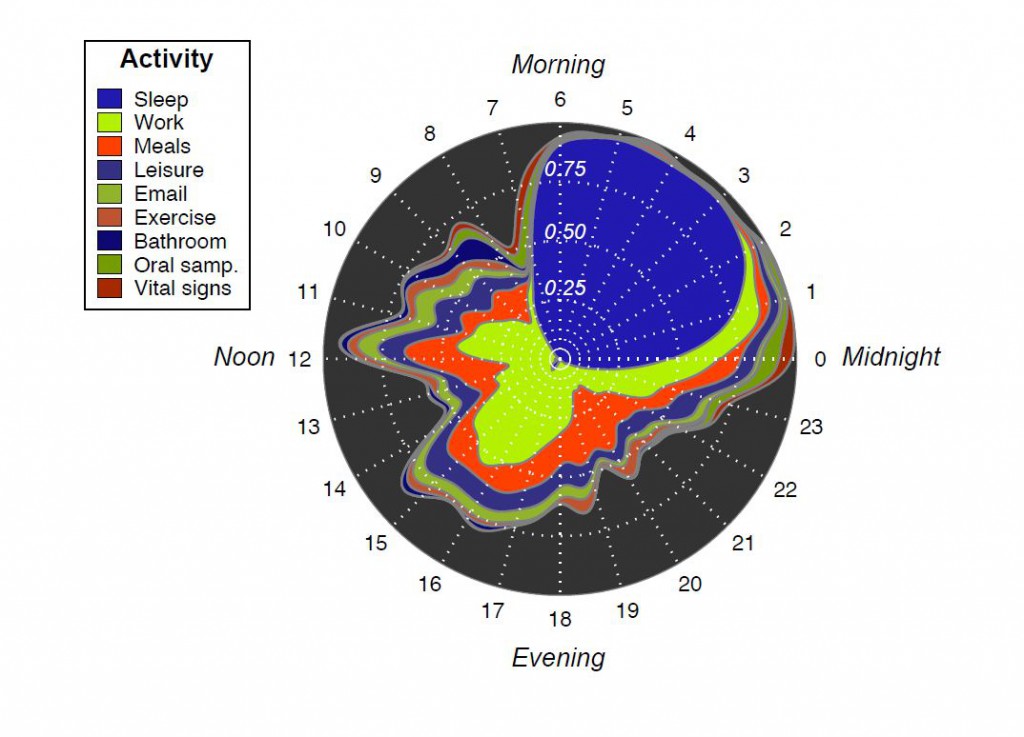
The aspect of this research that I find the most extraordinary is the data collection itself. Two individuals generated 10,000 observations of their diet, health, wellness, and behavior, comprising characterization of 349 different types of measurements a day, for a year! On top of that each individual also sampled their saliva and stool to characterize their microbial community.
How likely is it that any study could convince people to take such a diligent approach to self-recording and monitoring? Probably extremely unlikely. And yet, just as I did when I wanted to lose weight, millions of people around the world monitor aspects of their diet, exercise and activity with cell phone apps and online programs on a daily basis. As a result, we may be surprised how many people would want to parameterize their life in this way.
The study by Alm and colleagues is still only a study of two individuals, therefore, peoples self-interest in parameterizing their life will be essential if we are to expand this study to determine if the observed trends are repeated over hundreds to thousands of participants.
Of course, the time series aspects of this research is invaluable, as it enabled the researchers to demonstrate the remarkable stability of the microbiome, which has been shown in previous studies in Genome Biology (e.g. Moving Pictures of the Human Microbiome).
But it also enabled the detection of key events in the lives of these participants, including a bout of food poisoning, and the impact of travel to a developing nation on the microbial community.
This is extremely useful information, suggesting that the human microbiome can act as a forensic or diagnostic element of our human condition, with the potential to be able to determine not just disease burden, but lifestyle changes and even geography.
One of the key findings from this study is that fiber intake has a profound influence on the microbial assemblages of these individuals on a day-to-day basis. I have managed to maintain my weight loss, keeping at a healthy 165lbs, probably due to significant shifts in my diet, including fiber, and by virtue of maintaining an active lifestyle.
To monitor daily shifts in the microbial assemblage of my body would certainly be a valuable metric to measure, but I imagine that to do this for everyone would require a magic toilet and toothbrush that did it automatically. I would be willing to invest capital in such an idea.
I am very excited by the prospect of a frame shift in our health monitoring through these technological developments, though I will still likely find it hard to monitor 349 different variables on a daily basis!
Jack A. Gilbert
Latest posts by Jack A. Gilbert (see all)
- Microbial monitoring: health forensics for the modern age - 25th July 2014
Comments