![]() It seems that scientific research in the last two hundred years or so has made a full conceptual circle. In the good old days of the nineteenth century, any Englishman with a vaguely middle-class background, a source of modest income and an insatiable curiosity could treat research as an eccentric pastime. Whether that meant sea voyages and bird watching or hiking trips and rock collecting, what mattered first of all was the pursuit of the understanding of how the world works.
It seems that scientific research in the last two hundred years or so has made a full conceptual circle. In the good old days of the nineteenth century, any Englishman with a vaguely middle-class background, a source of modest income and an insatiable curiosity could treat research as an eccentric pastime. Whether that meant sea voyages and bird watching or hiking trips and rock collecting, what mattered first of all was the pursuit of the understanding of how the world works.
But times changed, and the world changed, and research moved from gentleman’s clubs to academic institutions. And although it gained more structure, and although, as hobbyists turned into professionals, scientific dillydallying turned into a rigorous process, we lost something important: the ability of the every(wo)man to satisfy his or her inquisitive mind through participation in the research process. And the more that was discovered, the weaker the connection became between academia (and research, and professional science) and the lay public.
The power of the crowd
One of the first to realize that the potential of the everyman could be put to better use were the brave men and women going where no man has gone before: the researchers of the SETI program, which searches cosmic radiowaves for signs of extraterrestrial life. Although the data analysis performed as part of the program used a lot of computing time, it did not require powerful machines: most operations could be done on simple personal computers. In the late 90s, the SETI@home project was started: users from all over the world were encouraged to download and install a small application, which would dip into the idle time of their computers to analyze chunks of SETI data.
Although no signs of extraterrestrial life have been found so far, SETI’s approach of using the power of crowd computing has become increasingly popular (in the last few years similar platforms called MilkyWay@home and Einstein@home were set up). Importantly though, SETI did not so much make use of the power of the crowd as it did the power of the crowd’s computers. The former, in which the pooled human power of the crowd is utilized, came later; and one of the best known examples is Foldit.
Foldit was developed from one of the SETI-inspired crowd computing projects, Rosetta@home. It is a science game in which the players, from whom no scientific background is requested, search for the true folded shape of a protein. Solving the shape of a protein has huge ramifications for, for instance, drug design. But any given amino acid chain can be folded in a myriad of different ways (a problem that must have given Levinthal a hell of a headache), and only a few, or even just one, of them represent the shape which we encounter in nature. While computers these days do a pretty decent job of finding a good approximation of these ‘natural’ shapes, they often miss out on details that can be perfected by the puzzle-solving players of Foldit.
Foldit offers users something that SETI@home didn’t: it makes them feel like true participants of scientific research, contributing not only the equipment (or computing power), but also their skills and, with time, even experience. And the recognition isn’t just nominal – in the first publication, which appeared in 2010 in Nature, and which described how the potential of online gamers can be utilized for biological research, Foldit players (as a collective) were listed among the article authors.
Harvesting the collective
Using the resources, time and skills of the public has undergone a boom in the last decade. SETI@home and Foldit represent two types of contribution: crowd computing and gamer-contributed system optimization. Another whole class of projects seeks public help with classification. This started in the mid-2000s with a project which originated from one Oxford grad student’s attempt to finish his PhD: Kevin Schawinski (now at ETHZ) tried to identify elliptical galaxies from photos collected by the Sloan Digital Sky Survey. He quickly figured out that much more could be achieved when galaxy classification was outsourced to the always eager internet crowd. Thus, just over half a decade ago, the Galaxy Zoo was born.
Galaxy Zoo can easily be called a tremendous success. Not only because of the incredible efficiency of its participants (the initial goal of 1 million classified galaxies was achieved within a month; before that Schawinski managed to classify a mere 50,000 in a week of ‘mind-numbing work’), and not only because of the sheer number of participants drawn to the project, but also because members of the lay public, who often had little idea of advanced astronomy, managed to make meaningful contributions to science – a spectacular example of this being the discovery of Green Pea galaxies. Today, building on the success of Galaxy Zoo, a huge number of other classification projects have been launched. They are collected under the umbrella of Zooniverse and range from other astronomical programs (for example, hunting for exoplanets), through climate sciences (classification of tropical cyclone data) and zoology (categorizing sounds that whales make or identifying African animals in photos from trap images), to medicine (analysis of cancer samples).
Projects and programs are defined as citizen science irrespective of the actual degree of public contribution. And although the allure of citizen science lies in its ‘democratic’ character, it is not just another sexy name for an empty concept. Quite the opposite: citizen science is strongly rooted in the idea of collective intelligence (understood in terms of Surowiecki’s four criteria, rather than consensus decision-making). In short, given a group of independent people of diverse experiences and opinions, the combined judgement they make as a group is better than any judgement made by an individual member. In this framework, the only thing that is needed to harvest collective intelligence is the tool to turn private assessments of all members into a collective decision of the whole group. In citizen science, these tools are science games.
Realigning the paradigm
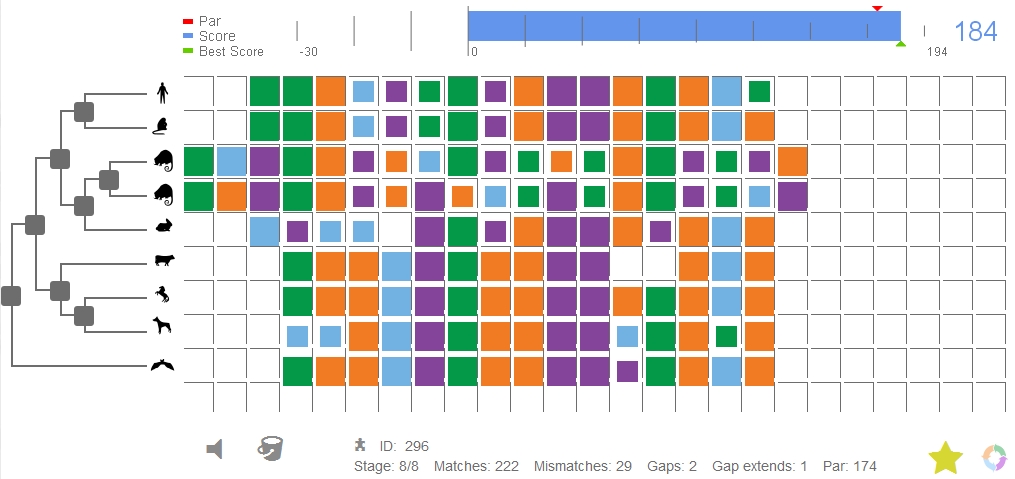
This week, Genome Biology publishes an article by Prof Jérôme Waldispühl and colleagues which makes an interesting contribution to the field of citizen science. Open-Phylo builds on the authors’ previous platform, Phylo, which was designed to solve multiple sequence alignment (MSA) problems.
Gamers trying to solve MSA puzzles in Open-Phylo do not have to have any scientific background. There are only a few simple rules that they need to be aware of (this knowledge can even be acquired simply by experience of playing the game): more squares of the same color (representing sequence similarity) in a column results in a better alignment; the similarity of neighbouring sequences counts for more than that of distant ones (i.e. better alignment between closely related species is prioritized over similarity of distant species); and the fewer gaps introduced into the sequence, the better. Armed with these rules, the players can go and align away. And, perhaps unsurprisingly, the alignments they achieve – as the authors of the article show – are better than those achieved by alignment algorithms, echoing the findings of Foldit.
One thing needs to be said here to avoid being cruelly unfair to the many talented creators of alignment programs: in Open-Phylo (as in all other optimization science games), the initial alignment is made by a computer. This is because the existing algorithms are already very good and, as with Foldit, are capable of obtaining quite satisfactory results. As it turns out, however, in every task performed by a machine, there is still space for a human intuition-driven improvement.
But Open-Phylo isn’t just yet another optimization science game. In a way unique to the world of citizen science and science games, it breaks the ground and shifts the paradigms. ‘How?’, you may ask. While most science games and citizen science projects are open to everyone at their output end, i.e. anyone can try to solve the puzzles, most of them have fixed input: the problems available for solving are strictly defined and provided by the platform authors.
Open-Phylo, however, allows any researcher to upload their problem to the platform to be solved by its players. In other words, it transforms the citizen science tool from a platform which simply collects individual assessments into a platform which directly connects researchers who have a specific MSA problem with internet folk and their crowd wisdom. And we should not be surprised to see other citizen science projects going in the same direction in the near future.
2 Comments