![]() Is biology at its boldest in life's nooks and crannies? Granted, Earth's most exotic organisms might not be quite so flamboyant as to have replaced phosphate with arsenate, as was momentarily suggested. But have a rummage through the annals of BioMed Central, and you will find that tales of the unexpected are most frequently to be found in little‑studied species and viruses. Permuted and split tRNAs in obscure Archaea? Check. A hybrid genome derived from RNA and DNA antecedents, in a virus so obscure that its identity is unknown? Check.
Is biology at its boldest in life's nooks and crannies? Granted, Earth's most exotic organisms might not be quite so flamboyant as to have replaced phosphate with arsenate, as was momentarily suggested. But have a rummage through the annals of BioMed Central, and you will find that tales of the unexpected are most frequently to be found in little‑studied species and viruses. Permuted and split tRNAs in obscure Archaea? Check. A hybrid genome derived from RNA and DNA antecedents, in a virus so obscure that its identity is unknown? Check.
Two new articles published today in Genome Biology's special issue on epigenomics once again remind us that biology loves to break all the rules, as they reveal the presence of DNA methylation in very surprising places. One article shatters the assumption that DNA methylation is absent in nematodes, while the other, astonishingly, finds methylation in a species with no characterized DNA methyltransferase – an enzyme thought to be essential for generating methylated DNA.
Never write off a nematode
C. elegans, science's most studied nematode, is a bona fide methylation-free zone – a curious exception to the widespread presence of DNA methylation across the tree of life. By extension, conventional thinking has held that DNA methylation is missing from all nematodes…until now, as researchers studying the parasitic worm Trichinella spiralis describe, for the first time, the presence of both DNA methylation and a DNA methyltransferase in a nematode.
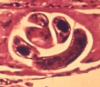 T. spiralis is one of the most widespread meat-borne parasites; it infects a broad range of animals, including humans, and causes trichinosis. The discovery of DNA methylation in this worm has an especially exciting twist, in that genes marked by methylation include those related to parasitism, making the study's authors hopeful that a new avenue for therapeutics research has been opened. A second striking observation: DNA methylation is incredibly specific to the parasite's life cycle stage – T. spiralis cycles between new-born larval, encysted muscle larval and adult stages – with zero DNA methylation in new borns increasing to a maximum in adults.
T. spiralis is one of the most widespread meat-borne parasites; it infects a broad range of animals, including humans, and causes trichinosis. The discovery of DNA methylation in this worm has an especially exciting twist, in that genes marked by methylation include those related to parasitism, making the study's authors hopeful that a new avenue for therapeutics research has been opened. A second striking observation: DNA methylation is incredibly specific to the parasite's life cycle stage – T. spiralis cycles between new-born larval, encysted muscle larval and adult stages – with zero DNA methylation in new borns increasing to a maximum in adults.
The mystery of the missing methyltransferase
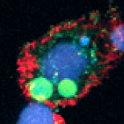 DNA methylation in T. spiralis can readily be explained by the presence of a (previously unnoticed) DNA methyltransferase – an enzyme that intriguingly has no known counterpart in any other nematode. But trickier to explain is the new report of DNA methylation in the ciliated protist Oxytricha trifallax, an organism that has no recognizable DNA methyltransferase gene.
DNA methylation in T. spiralis can readily be explained by the presence of a (previously unnoticed) DNA methyltransferase – an enzyme that intriguingly has no known counterpart in any other nematode. But trickier to explain is the new report of DNA methylation in the ciliated protist Oxytricha trifallax, an organism that has no recognizable DNA methyltransferase gene.
The peculiar two-genome set-up of ciliates, in which one genome is heavily edited and remixed to form a second genome (both residing in the same cell), makes reproduction a very complicated process for Oxytricha, involving DNA rearrangement and elimination. One of the functions of DNA methylation in Oxytricha seems to be to mark DNA sequences for destruction during reproduction, most likely in concert with non-coding RNAs. Intriguingly, hydroxymethylation (the so-called '6th base') also makes an unexpected appearance in Oxytricha.
The apparent absence of a known DNA methyltransferase in its genome makes Oxytricha a lonely soul: the only species in which the origin of its DNA methylation is a total mystery. The authors of the study controversially propose that a novel mechanism may be responsible for DNA methylation in these ciliates, and that it may be an ancient methylation machinery.
New insights need open data
The authors of our two exotic methylation studies did not come by their exciting findings easily, not least because a huge amount of data needed to be generated to confirm the controversial presence of DNA methylation, and to map its temporally dynamic location within the genome. The Oxytricha study includes high-throughput meDIP-seq data (for both DNA methylation and hydroxymethylation), bisulfite Sanger sequencing data for selected loci and mass spectrometry validation data. All these data are publicly available for download, as is detailed in the article.
 The nematode study is highly innovative in its approach to open data, with the supporting data (deposited in the GigaScience database) presented in the interoperable ISA-Tab format to maximize its reusability. These data include whole-genome bisulfite sequencing – the Rolls Royce of DNA methylation mapping protocols – as well as mass spectrometry validation data and bisulfite Sanger sequencing of selected loci. High-throughput data from the study have also been deposited in the standard NCBI databases.
The nematode study is highly innovative in its approach to open data, with the supporting data (deposited in the GigaScience database) presented in the interoperable ISA-Tab format to maximize its reusability. These data include whole-genome bisulfite sequencing – the Rolls Royce of DNA methylation mapping protocols – as well as mass spectrometry validation data and bisulfite Sanger sequencing of selected loci. High-throughput data from the study have also been deposited in the standard NCBI databases.
The value of open data for such breakthrough science lies not only in providing a resource, but also in conferring transparency to unexpected conclusions that others will naturally wish to challenge.
Further reading
Soojin V Yi: Birds do it, bees do, worms and ciliates do it too: DNA methylation from unexpected corners of the tree of life Genome Biol 13:174 (Research Highlight)
Scott Edmunds: First methylated nematode genome and other new datasets in GigaDB GigaScience blog
Beth Marie Mole: Epigenetic Enigmas The Scientist
Beth Marie Mole: Genomic Methylation Collector The Scientist
Latest posts by Naomi Attar (see all)
- tRFs and the Argonautes: gene silencing from antiquity - 2nd October 2014
- Keeping up with the Jobses: the role of technology in reproducible research - 26th September 2014
- How to disarm a superbug – a story told by forensic genomics - 23rd June 2014
[…] 18/10/12: See also the great Genome Biology blog posting and accompanying commentary on the nematode study. Tags: data, data citation, data publication, […]
[…] "A good kick in the rear" When The Scientist asked Paul Sternberg to comment on one of two special issue articles reporting the discovery of DNA methylation in unexpected phyla, his response was frank: "This is just a good kick in the rear to keep an open mind about all of these mechanisms, and to look harder for methylation in other species." To find out more about these articles, see either our accompanying Research Highlight or our blog post. […]
[…] articles set out to make all their raw data fully open, see Genome Biology’s blog post on ‘Rebel worms and mixed-up ciliates’. In recognition of these efforts, Fei Gao, first author of the T. spiralis article, recently […]