Schistosomiasis affects approximately 250million people across 78 countries. Control efforts have have reduced the prevalence and distribution of the disease globally, with the WHO outlining a goal of 2030 to eliminate schistosomiasis as a public health problem. The majority of schistosome infections (~90%) are reported in Africa, and are caused by infection with Schistosoma mansoni and S. haematobium. Schistosomiasis caused by infection with S. japonicum is, however, also widespread, with human infections reported across much of Southeast Asia, including China, The Philippines, and Indonesia.
Unlike African schistosomes, Schistosoma japonicum causes zoonotic schistosomiasis. As well as humans, around 40 species of wild and domesticated animals can be infected by S. japonicum, with animal hosts playing an important role in transmitting parasites to humans.
In China, attempts to control S. japonicum have been some of the most successful worldwide, reducing the number of human infections from 11 million to less than 100,000 today, as well as dramatically reducing the number of infected livestock.
End-game elimination and the need for a vaccine
To finally eliminate schistosomiasis in China, the development of an effective anti-schistosome vaccine is increasingly considered an essential tool in moving towards end-game elimination. A vaccine would also reduce reliance on mass drug administration of praziquantel, and help mitigate the possible evolution of resistance to the drug (covered in a recent BugBitten blog).
Over 100 vaccine candidate antigens have been tested against various schistosome species within animal models.
A particular protein family of interest, which have several members currently in vaccine trials, are the tetraspanins (TSPs) – a group of tegument-associated proteins that interact directly with the host’s immune system using their large extracellular loop (LEL) domains, and which have crucial functions related to schistosome transition to parasitism, survival in the definitive host, and have been implicated in helping the parasite evade immune recognition.
Vaccine studies using certain TSPs in S. japonicum (SjTSPs) have, however, shown variable levels of protection in animal models, highlighting variation in SjTSPs as possible causes of this variable efficacy.
Robust selection of target TSPs, including assessment of their genetic diversity and antigenic variability, is essential. Exactly how antigen diversity influences the antigenic properties of parasite antigens, such as SjTSPs, remains necessary to properly evaluate their application as vaccine targets.
A recent study published in Parasites & Vectors has used in silico bioinformatic and protein modelling approaches to attempt to bridge these knowledge gaps, and characterise the genetic diversity of SjTSPs from parasite populations in China. Seven SjTSPs (SjTSP-1, -2, -8, -13, -14, -23 and -25) were selected for analysis based upon published information on their structures, immunogenicity and vaccine candidacy.
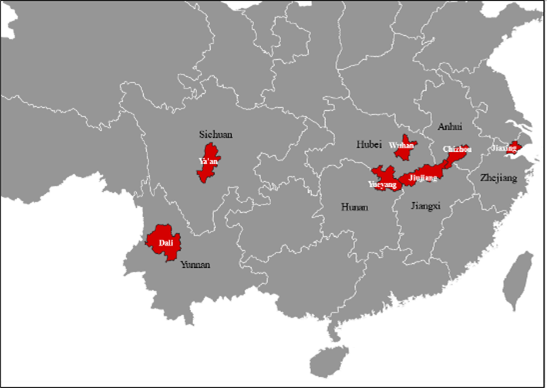
By searching S. japonicum genome sequencing data collected from parasites in seven Chinese provinces, and extracting SjTSP sequences from this genomic data, the geographic variability of the seven SjTSPs was assessed. The evolutionary selection pressures driving SjTSP geographical variation, and the impact of variation on the structures and immunogenicity of each SjTSP was also explored to evaluate their potential as vaccine targets, as well as mechanisms employed by the parasite to evade the host immune response.
Genetic diversity and sequence variability of TSPs between Schistosoma japonicum populations
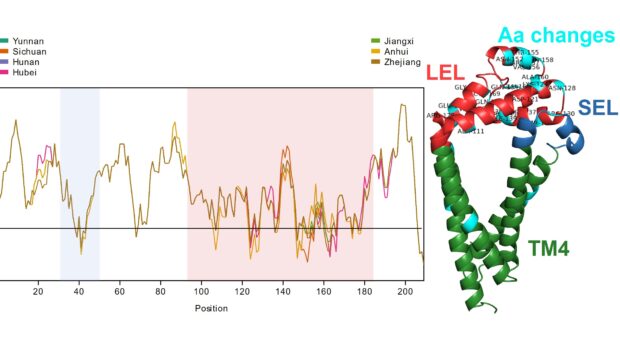
Considerable geographical variation was demonstrated between SjTSP-2, -8, -23 and -25 across the seven Chinese provinces. In particular, SjTSP-2 was shown to be the most variable, with 67 polymorphisms (i.e. mutations) identified between sequences representing parasites sampled from the seven provinces.
In contrast, SjTSP-1, -13 and -14 were comparatively more genetically conserved across the parasite populations, with very few polymorphisms identified.
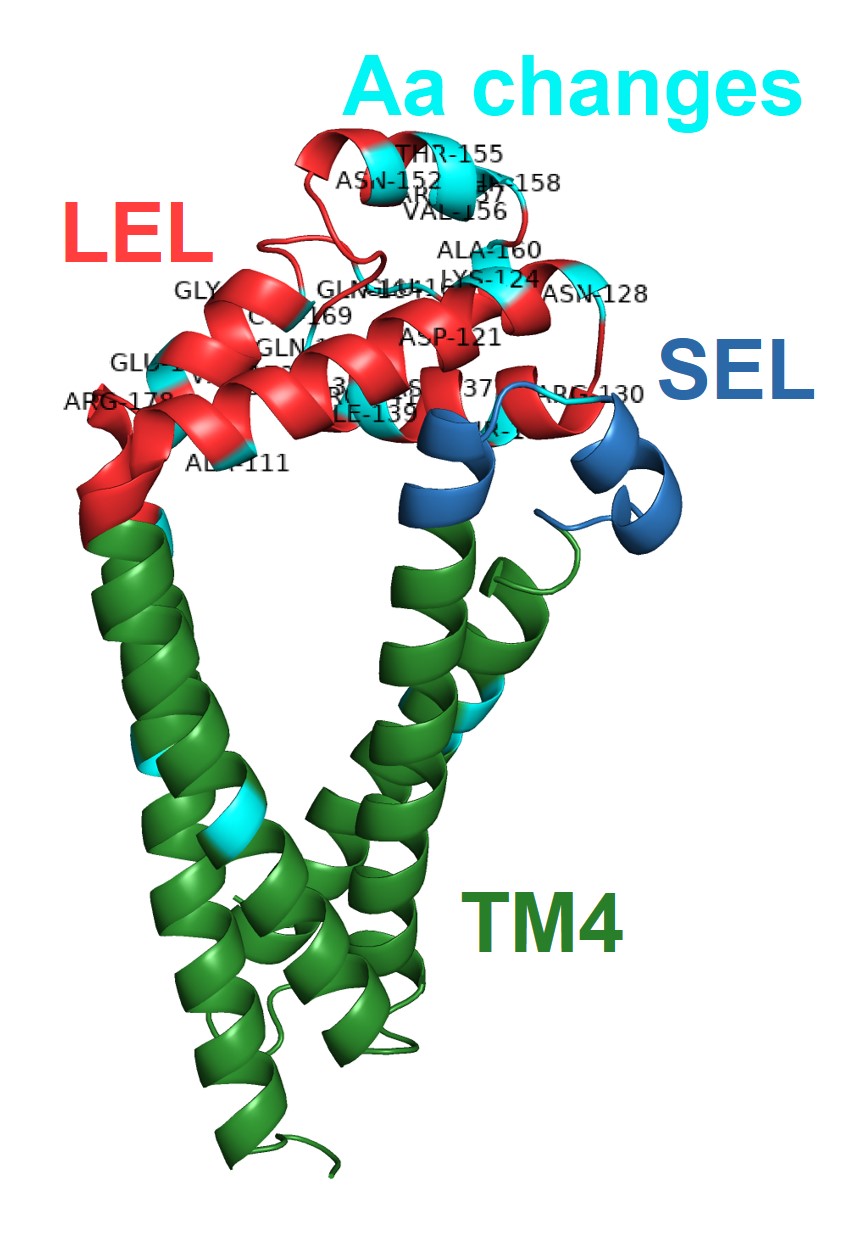
It was found that the majority of polymorphisms identified in the four variable SjTSPs resulted in the accumulation of amino acid changes in the functionally and immunologically important LEL domain.
In SjTSP-2 specifically, the extent of this LEL domain variation leads to the formation of a hypervariable region.
It was also predicted that selection pressures were acting on many of the same sites in the LEL domain where these polymorphisms were accumulating, suggesting that exposure of the LEL domain to the host’s immune system might be promoting variation between SjTSPs from parasites in different Chinese provinces.
Impact of genetic variation on SjTSP structures and antigenic properties
Overall, it was demonstrated that variation in genetically diverse SjTSPs from geographically distinct parasite populations resulted in considerable variation in their protein structures and antigenic properties. Across the whole antigen, the LEL domains of those variable SjTSPs, in particular, were found to contain the greatest fluctuations in antigenicity (i.e. likelihood of antibody binding).
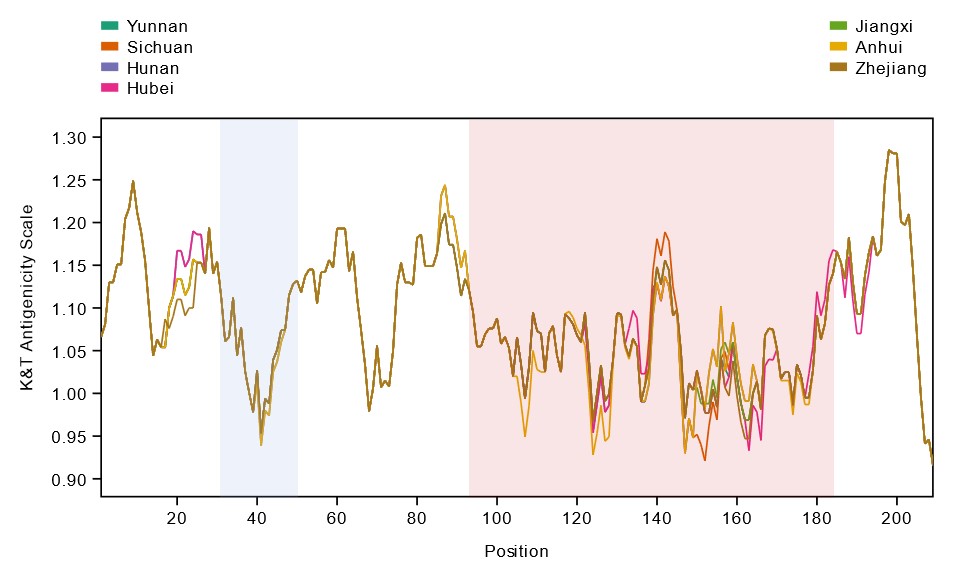
This variation in the structures and antigenic properties of these SjTSP LEL domains was generally predicted to limit antigen recognition by host antibodies, possibly providing SjTSPs with ‘escape’ mutations that facilitate evasion of the host’s immune response. Such changes seem to therefore represent a mechanism utilised by S. japonicum parasites to manipulate the immune system of their hosts.
On the other hand, limited genetic diversity and geographical variation of SjTSP-1, -13 and -14 results in a lack of both structural and antigenic variability.
Implications for SjTSP vaccine candidacy
This research shows that SjTSPs are attractive targets for a host immune response, and that selection pressures drive polymorphisms in the LEL domain of certain SjTSPs. This results in structural and antibody-binding changes in these antigens, and could feasibly influence the effectiveness of the host immune response.
Using these variable SjTSPs as vaccine targets could therefore represent a challenge for vaccine development against S. japonicum, supporting other findings that SjTSP variability is, at least in part, responsible for the variable protective efficacy demonstrated so far in vaccine studies.
If the variability observed amongst certain SjTSPs may reduce their suitability as vaccine targets, then conservation of SjTSP-1, -13 and -14 may highlight their potential as effective vaccine targets.
Robust evaluation of vaccine target genetic and antigenic variability in natural parasite populations is recommended, and should be a prerequisite for future vaccine development. This work also provides a possible analytical framework for the investigation of future schistosome antigen coding-genes as vaccine candidate antigens.

Comments