Multiple sclerosis (MS) is a chronic neurologic disease with known genetic influences. Genetics determine who acquires MS, how severe the disease will be and how quickly it will progress. MS patients show variable response to drugs with some patients showing significantly better responses than others. The MS research community is increasingly converging on the idea that the patients’ genetic profiles influence their response to MS drugs.
Copaxone is a drug that was discovered serendipitously as being effective against MS. While its clinical efficacy and safety are well-established in long-term studies and in clinical practice, its mechanism of action continues to be a subject of study. Research indicates that Copaxone’s protective effects against MS are, in part, due to its complex interactions with the immune system. Like other MS drugs, the drug-response to Copaxone varies from one patient to another.
In the May 2017 issue of Genome Medicine, my coauthors and I describe our investigation of the genetic basis of response variability to Copaxone in the largest pharmacogenetics study ever performed in MS. In 2,422 patients enrolled in ten clinical trials that collectively recruited from 553 clinical centers across 34 countries, we investigated the association between response to Copaxone and single nucleotide polymorphisms (SNPs), which are nucleotides on chromosomes that can exist in more than one form (i.e. are polymorphic) in the population.
Some of these trials include patients who have been continuously followed for more than 20 years, making our study one of the very few to examine response to an MS drug over such a long period of time. We discovered SNPs associated with Copaxone response in patient cohorts from two of the clinical trials and validated our findings in the remaining eight trials.
We found that a genetic signature comprising four SNPs had the greatest influence on response to Copaxone. Furthermore, the signature was not associated with response in patients who were treated with placebo or IFN-β (Avonex), another MS drug used in the trials. This indicated that the signature comprising the four SNPs is specific to Copaxone response and not simply an indicator of MS progression or of response to IFN-β. The SNPs are part of four genes, MBP, HLA-DQB2, UVRAG and ZAK (CDCA7).
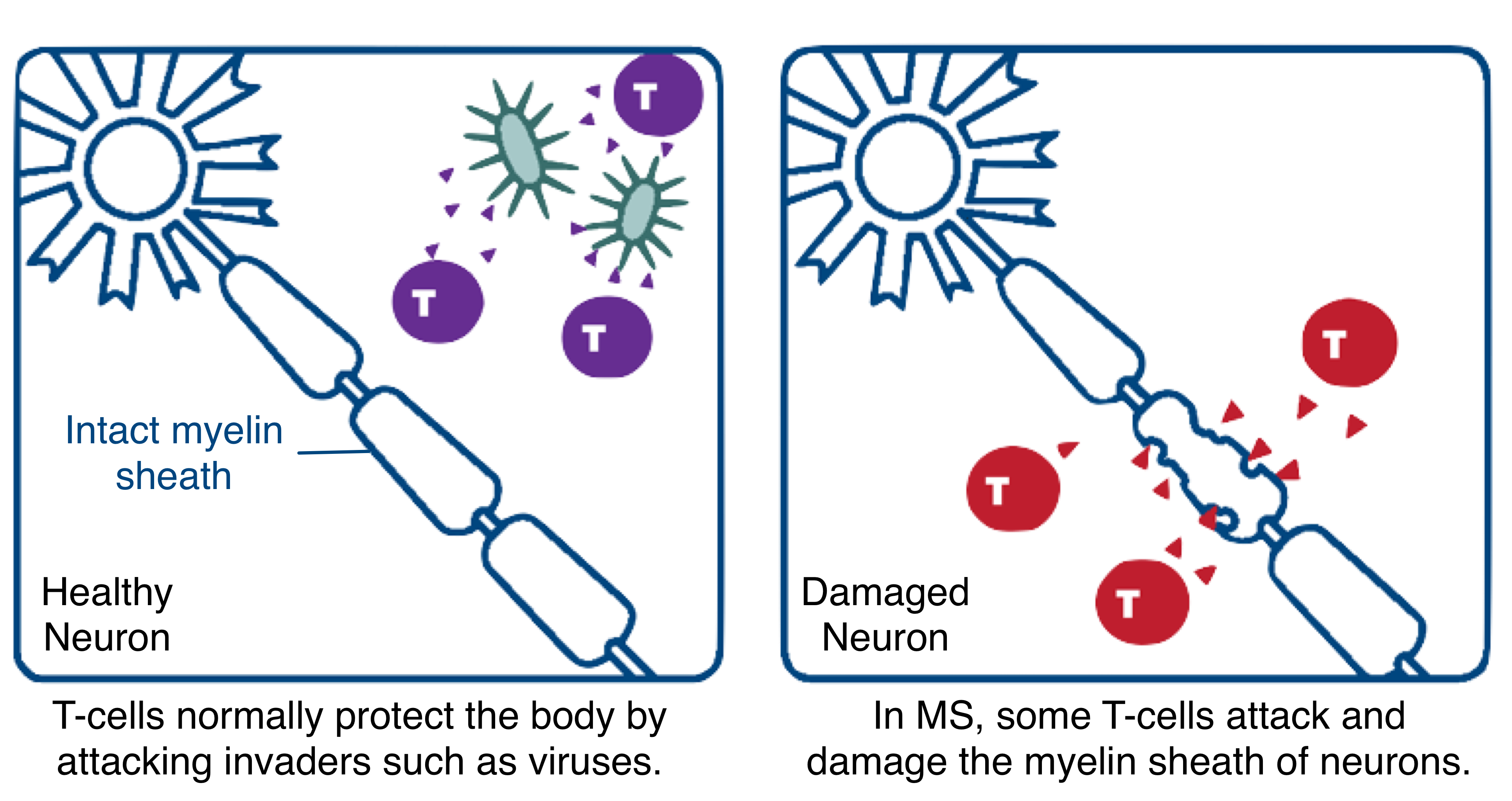
In the body’s central nervous system, the protective covering of the nerves, known as the myelin sheath, facilitates the rapid conduction of signals that enables cells to communicate with each other. In MS patients, misguided T-cells from the immune system attack the myelin sheath, stripping it away and damaging the nerves over time. HLA-DQB2 is a gene that produces proteins which enable the immune system to identify and then attack the myelin sheath.
By mimicking myelin basic protein (MBP), the protein that makes up the myelin sheath, Copaxone protects the sheath by lessening and diverting the impact of the immune system. UVRAG has recently been shown to influence the fate of T-cells in the immune system and ZAK is known to be activated during stress and is linked to brain lesions in MS patients. The biological roles of these four genes underlying the SNPs in the signature helped improve our understanding of Copaxone’s mechanism of action.
The four-SNP signature identified a ~10% subset of the MS patient population who experienced a 93% reduction in their annualized relapse rate compared to pre-trial rates. A similar observation was recently made by another group of researchers who employed SNP signatures to identify a 10% subset of patients who did not respond to IFN-β, another major MS therapy.
These findings indicate that the effect of each SNP on the drug-response is small. Even when multiple SNPs are combined in a single signature, they are unable to explain the drug-response in many patients and are not powerful enough for clinical applications. This emphasizes the need for studying the effects of many SNPs together, and not one-by-one, a strategy that is easier and often employed when there are several SNPs to analyze.
The widely variable clinical definition of what constitutes a drug-response in MS also presents a challenge. We found that the four-SNP signature, which was discovered using one of the clinical definitions of drug-response, was in fact, consistent across many other commonly used definitions of drug-response.
The findings in this study are an encouraging start for identifying genetic signatures that can help guide MS clinicians to select the best treatment options for their patients. Nevertheless, as a research community, we have a long way to go to fully understand which patients will respond best to any given drug, including Copaxone.
Comments