Although genome sequencing has greatly aided marker-assisted breeding, it still takes a number of generations to produce crops with altered traits. Conventional genetic modification allows for individual or multiple genes to be transformed into crops. A recent exemplar of this is Golden Rice. However in many crop plants, limitations with transformation efficiency and also the current regulatory environment make the use of transgenic technology more onerous than ordinarily envisioned.
Genome editing has the potential to overcome these limitations by precisely targeting genes of interest in early generations. In addition, as ‘gene editor’ proteins can then be crossed out of your crop, it leaves no ‘transgenic’ residue and should, in theory, be subject to different regulations to conventional genetically modified. This is the current opinion of the BBSRC.
Broadly speaking, genome editing technologies rely on molecular mechanisms that precisely target specific DNA sequences and then use nuclease activity to cut those sequences. The target cells’ DNA-repair mechanisms will then ‘fix’ this mistake, but will often do this imperfectly, introducing mutations that alter the activity of the subsequently produced protein.
There are three broad types of genome editing that are being used in plants, namely zinc finger nucleases (ZFN), transcription activator–like effector nucleases (TALENs) or the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated system (CAS). ZFNs are comprised of a DNA binding sequence (the zinc finger) linked to a nuclease domain that cuts the DNA.
ZFNs have been used in a variety of organisms but their molecular construction can be non-trivial and therefore more difficult to use than other technologies.
TALENs use a bacterial system that evolved to target expression of host genes by precisely binding to host DNA sequences. When this targeting-protein is linked to a nuclease, precise cuts can be made at a specified sequence. Although this technique has been widely used in animals, to date there have been few examples of its use in plants.
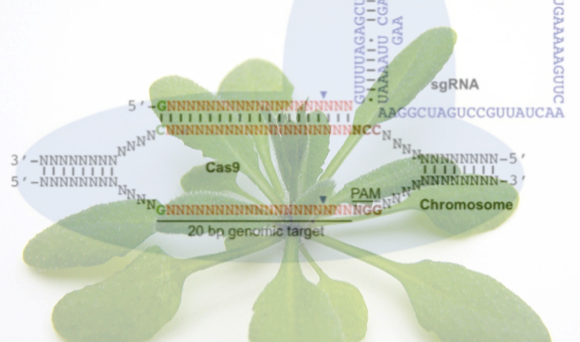
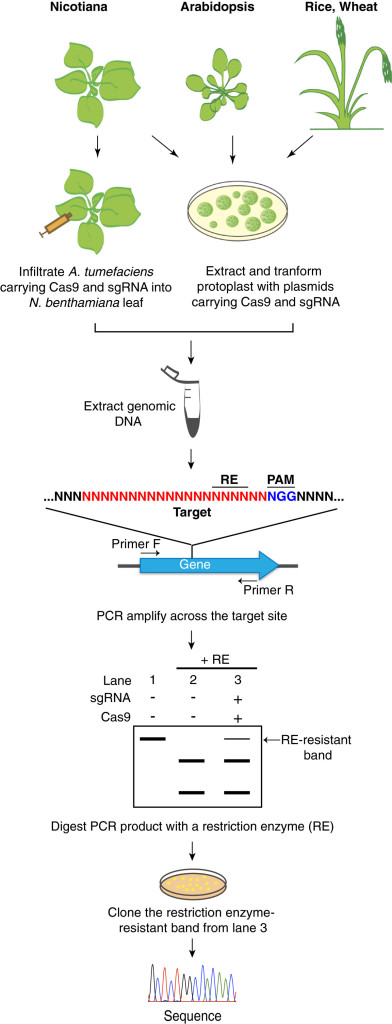
The latest and most popular genome editing technology is the CRISPR-Cas system. This is based on an adaptive bacterial immune response and relies on a single Cas9 nuclease targeting a specific sequence defined by a single guide RNA (sgRNA). The ease of changing the specificity of the nuclease by altering the sgRNA makes this an attractive option for researchers. Unfortunately this system also includes a certain amount of tolerance to mismatches between the sgRNA and the target DNA sequence so can be prone to generation of off-target mutations, which might lead to unintended phenotypic consequences.
Use of this system requires a high level of quality control, to ensure that only the target sequences have been mutated. However, as knowledge of the system increases it will improve our ability to guarantee precise targeting. Another benefit of the CRISPR-Cas system comes in the ability to target gene families that share the 20mer sgRNA sequence. In theory this will mean that a single sgRNA-Cas9 transformation could target an entire family of genes.
Although each of these genome editing technologies holds great promise as methods of crop improvement, many technical aspects have yet to be optimized.
 For this reason it is an ideal time for Plant Methods to support a thematic series on ‘Plant Genome Editing’, to review the current state of the field and to highlight recent advances in this technology.
For this reason it is an ideal time for Plant Methods to support a thematic series on ‘Plant Genome Editing’, to review the current state of the field and to highlight recent advances in this technology.
 This is timed to coincide with a CRISPR workshop jointly organized by the GARNet and OpenPlant networks, supported by Plant Methods and bringing world experts together to introduce and discuss the technology with those interested in adopting it in their own research programs.
This is timed to coincide with a CRISPR workshop jointly organized by the GARNet and OpenPlant networks, supported by Plant Methods and bringing world experts together to introduce and discuss the technology with those interested in adopting it in their own research programs.
 The deadline for applications is Monday August 10th and registration details can be found here.
The deadline for applications is Monday August 10th and registration details can be found here.
Geraint Parry
Latest posts by Geraint Parry (see all)
- Genome editing in plants - 7th August 2015
Comments