The reliability and reproducibility of scientific data are subject to increasing scrutiny in the light of concerns raised in particular about preclinical research, but are also much more widely acknowledged now. Some of the issues are simple to identify, if not necessarily to address – choosing the right sample size, applying appropriate statistics, identifying reagents and other resources.
Others may be more subtle, and one such example has emerged from recent close examination of the effects of reagents widely used to explore gene function. Different approaches that claim to answer the same question may, it turns out, deliver different answers – but for reasons that couldn’t have been predicted.
Down or out?
The time-honored approach to finding the role of a gene is to interfere with its function. There are two broad ways to do this: knockout mutations that remove the gene, or knockdown, which reduces expression of the gene product with interference (antisense) technology.
In principle, these should have the same effect — because the protein product is absent or disrupted and thus non-functional; and because of its low cost and speed, morpholino knockdown technology based on antisense has been widely used.
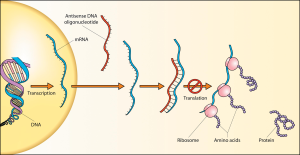
But for some time, concerns have been growing about the failure of morpholino knockdowns to show the same phenotype as knockouts, with a paper from Nathan Lawson’s group suggesting that around 80% of phenotypes seen using morpholino knockdown technology in zebrafish were not seen in mutants of the same genes.
So are null mutants the ‘purest’ way to find a gene’s function, and knockdowns unreliable? There are indeed known hazards with knockdown approaches: off-target effects, where the interference machinery affects genes other than the intended target; overloading the interference machinery; and inducing other confounding effects too byzantine to specify here.
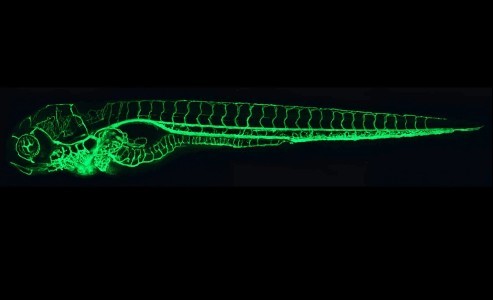
Now, work from Didier Stainier’s lab shows a much more interesting underlying issue. The group focused on the egfl7 gene, because while mouse mutants show no obvious phenotype, knockdown in a range of organisms leads to severe vascular defects.
They first confirmed this strong phenotype in zebrafish when using morpholino-mediated knockdown of egfl7, but as with mice, a strong mutant allele of egfl7 did not show any gross effects.
Were the knockdown phenotypes due to off-target effects? If the morpholino was affecting other targets, then its use would enhance the otherwise mild mutant phenotype. If, however, the morpholino was truly specific, then it should not have additional effects on the mutant, since the gene would already be inactive and there would be nothing for the morpholino to interfere with.
The group showed that the morpholino indeed had nothing to add to the mutant phenotype – and also ruled out a range of other potential explanations. Why, then, the difference in effects?
Redundancy and robustness
The answer seems to be genetic compensation. Two proteins were strongly upregulated in the egfl7 mutant, but not in the knockdown, and artificial introduction of either Emilin2 or Emilin3 mRNA rescued the knockdown phenotype.
It’s not clear exactly how this compensation works, but it appears to be activated only by deleterious genetic lesions, and not by interfering with the expression of gene products. A similar phenomenon was also seen with another gene, suggesting that this may be common.
Genetic redundancy, where two or more genes can encode a similar biochemical function, is an important feature of robust biological networks (although of course it can also notoriously make it difficult for researchers to determine the precise role for the genes involved). Now it seems that mechanisms also exist for preserving functional integrity of a biological system post-transcriptionally.
The right tool for the job
The trick is to try to be sure about what question you want to ask, and what question a particular method is actually going to help answer.
The work also raises the question of what we’re looking for when we investigate the ‘function’ of a gene: is the knockdown the ‘true’ phenotype, which in this case shows that egfl7 is a critical vascular gene, or is the mutant ‘true’, and more representative of biological reality, and deletion of egfl7 is not so important for the organism?
Finally, returning to the theme of reproducibility in research, both methods might be reliable (repeatable) in the results observed in this case; the trick is to try to be sure about what question you want to ask, and what question a particular method is actually going to help answer.
Judging that might be harder than, for instance, validating that a cell line really is what it’s supposed to be (one of the more recent issues in the general reproducibility debate), but seems just as important to consider when designing and interpreting experiments.
Liked the blog? Read the original paper by Rossi et al.
Graham Bell
Latest posts by Graham Bell (see all)
- Naked mole-rats, scientific villages and the Oscars of science - 15th December 2017
- Non-model models, preprints, and the fruit of the translational tree - 9th December 2016
- What is wrong with this picture? – reproducibility and realism - 23rd June 2016
Comments