Synthetic Genomics: Redesign and synthesis of the first multicellular eukaryotic genome
The field of synthetic biology, designing and building engineered biological systems through DNA synthesis and genetic engineering, is rapidly moving to a genome scale. In a similar trajectory to genomic sequencing and genome projects two decades ago, it has moved from engineering single genes, entire synthetic bacterial genomes (J Craig Venter’s notorious “Synthia”), to the eukaryotic organism stage. The “Sc2.0” synthetic yeast genome project, is an international consortium synthesizing “designer eukaryotic genomes“ for all of the 16 chromosomes (and roughly 14Mb of sequence) of Baker’s yeast Saccharomyces cerevisiae. With the first chromosome published last year in Science, and many more already completed, and planning for the next step up is already being discussed. Being heavily involved in Sc2.0, and already producing several chromosomes (pictured), our colleagues at BGI organized a workshop covering this very topic at the end of last year. Chantal Shen and Huanming Yang from BGI and Patrick Cai from Edinburgh have written us a guest posting on the discussions that went on at the workshop, and what the proposed next steps are for organismal scale synthetic genomics.
The first workshop to discuss and propose the international collaboration on the projects in the field of synthetic genomics after the Sc2.0 project was held on Sept. 12 in Shenzhen, China, as a part of BGI’s 9th International Conference on Genomics (for a write up of ICG9 see this report from BGI). The workshop was attended by the participants of the Sc2.0 community from the USA, Europe, and China, as well as their funding agencies.
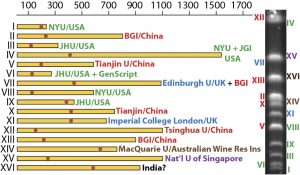 The publication on the synthesis of Chromosome 3 of the yeast genome on 28th March 2014 indicates the successful strategic roadmap and technical protocol of the Sc2.0 project. Groups responsible for all other yeast chromosomes have made good and smooth progresses. It is reasonable to be optimistic about completion of all chromosomes ahead of schedule, even if the final integration into a whole genome still remains a serious challenge. While work is progressing rapidly, now is the right time to discuss what the international participants of the Sc2.0 consortium should do next.
The publication on the synthesis of Chromosome 3 of the yeast genome on 28th March 2014 indicates the successful strategic roadmap and technical protocol of the Sc2.0 project. Groups responsible for all other yeast chromosomes have made good and smooth progresses. It is reasonable to be optimistic about completion of all chromosomes ahead of schedule, even if the final integration into a whole genome still remains a serious challenge. While work is progressing rapidly, now is the right time to discuss what the international participants of the Sc2.0 consortium should do next.
The workshop discussed two options in the coming years: 1) to synthesize another more complicated monocellular genome with high throughput, automated techniques, e.g. fission yeast or other higher fungus-like mushrooms, to synthesize another monocellular eukaryotic organism with the differentiation-like phenotypes, or to emphasize more on the application to yeast breeding. The main argument for this approach was to further encourage the positive responses from the public on synthetic biology and anticipate further innovative technologies to further lower the costs by integrated automation platforms; 2) to go directly to the synthesis of the first multicellular eukaryotic genome.
The workshop reached the consensus that the next strategic mission for the international consortium was to move towards the redesign and synthesis of the first multicellular eukaryotic genome, while other options as above, which should not be excluded, could be done by individual labs. This was thought to be the most natural development post-Sc2.0.
The workshop included seriously discussion and debate on the technological challenges, emphasizing that the strategy of “genome replacement” or homologous recombination might not be wholly applied to multicellular organisms. The refined nuclear transfer technology for the whole genome synthesis approach, and any other relevant emerging technologies, such as CRISPR (see the recent review in GigaScience), should also be considered. The collaborative approach with other communities, such as those in biology, is essential. To be practical, the cost of synthesis, which is approximately 10c per base now (one tenth of what it was at the beginning of the Sc2.0), will need to further fall to one cent per base in 5 years, or even lower.
The workshop also discussed and debated on the choice of the candidate genome. The criteria for choice might be: 1) a smaller genome size; 2) a well sequenced and annotated reference genome, 3) being well characterized biologically and physiologically, 4) well established culturing and phenotyping techniques.
 The road ahead: synthetic worms and beyond?
The road ahead: synthetic worms and beyond?
As strongly suggested by Sydney Brenner (pictured visiting us at BGI) and his colleagues in the nematode field, C. elegans, with a genome size approximately 6 times larger than yeast (100Mb), and amongst the best characterized model organisms in terms of its biology and development, was considered as one of the strongest candidates, although other model plant (e.g. Arabidopsis) or animal (e.g. Drosophila) genomes were also looked at in the workshop. The differences among the different candidates, both the advantages and disadvantages to our current knowledge were compared. In addition to this, synthesizing an intact or a part of a chromosome of a higher eukaryotic organism was also proposed. The final choice will continue to be to be debated before the next meeting.
The workshop has proposed the general timetable for this one-decade-initiative between 2015-2024: 2 years for discussion, 3 years for pilot experiments, and 5 years for large-scale implementation. As with the Human Genome Project, the participants felt there was a high likelihood of the project to be initiated and completed sooner than even this speedy timeline due to the continuing rapid pace technological breakthroughs, if managing to avoid any hesitations and delays. The participants pledged their responsibility to carry the work out once the tasks were assigned.
The workshop called for the broader international discussion about the choice of the candidate genome, the strategy and details of the technology, the roadmap, protocol and timetable of the initiative. Wider participation and discussion from the various communities working on the biology of the candidate organisms, and technical innovation and development, was called for.
The principles of international collaboration and open data/tech-sharing were again reaffirmed for the follow up project. As with China participating in the Human Genome Project, groups from other developing countries were encouraged to join and share this opportunity to contribute to another milestone in the life sciences. However, the facilities and capacities to guarantee its completion according to the schedule would need to be evaluated.
The workshop also suggested the setting up of a coordination committee and a 3B (bioethics, biosafety, and biosecurity) group to initiate the relevant discussions. “From scratch” approaches and “do-it-yourself” advocates were also considered. The participants also proposed the set up of a “SynBio Code” for all the future members of the international synbio community.
The next workshop or strategic meeting will be held in May 2015. The meeting location is still to be decided but will be chosen in a competitive way. Watch this space for further news on how these projects are continuing.
Huanming Yang, Chairman, BGI Shenzhen
Patrick Cai, Director, Edinburgh Genome Foundry
Chantal Shen, Unit of Synthetic Biology, BGI Shenzhen