The difficulties sharing neuroscience data: can data publishing help?
 Last week we published our first neuroscience data note containing 10GB of fMRI data hosted and integrated into the paper by a DOI to our GigaDB database. While we have published a number of genomics datasets and data notes (see the Puerto Rican Parrot genome data note and its associated data DOI), this is a nice example of us providing a home for “orphan data”, the long tail of data types without community agreed curated repositories. Sharing of data enables re-use and new work to be created, all goals and reasons why we have built the infrastructure that makes up GigaScience and its integrated data hosting environment. This is even more timely, with a number of recent and high profile retractions and fraud cases in the areas such as psychology and animal cognition showing there is a growing need for increased transparency and access to supporting data to tackle this growing reproducibility gap.
Last week we published our first neuroscience data note containing 10GB of fMRI data hosted and integrated into the paper by a DOI to our GigaDB database. While we have published a number of genomics datasets and data notes (see the Puerto Rican Parrot genome data note and its associated data DOI), this is a nice example of us providing a home for “orphan data”, the long tail of data types without community agreed curated repositories. Sharing of data enables re-use and new work to be created, all goals and reasons why we have built the infrastructure that makes up GigaScience and its integrated data hosting environment. This is even more timely, with a number of recent and high profile retractions and fraud cases in the areas such as psychology and animal cognition showing there is a growing need for increased transparency and access to supporting data to tackle this growing reproducibility gap.
The challenges sharing neuroscience data
Genomics has been a fertile area for data sharing through having a relatively limited number of platforms and resulting data types, and being unified by being focused on a particular technology. This has made it very different to an area such as neuroscience that falls into separate communities, each studying the brain at different levels and with different tools, which has made it much harder to provide motivation and a need to provide data to researchers outside their community. Compared to the (relatively) smooth rise of genomics data sharing led by the Human genome project and community guidelines formulated at the Bermuda and Fort Lauderdale meetings, moves to spur similar efforts in neuroscience have been more challenging. Organizations such the International Neuroinformatics Coordinating Facility (INCF) have been set up to coordinate and encourage neuroimaging data sharing, and have been producing tools and infrastructure to enable it.
One of the many tools promoted by the INCF has been the fMRI data center (fMRIDC) — a large-scale effort to gather, curate, and openly share fMRI data used in peer reviewed studies. Pioneering data sharing in neuroscience, despite receiving support from journals such as the Journal of Cognitive Neuroscience, the platform initially received skepticism from many of the community due to concerns over scooping and practicalities such as the time and effort it would require to deposit usefully curated data. The resource helped prove its utility over the following years by collecting over 100 complete studies, and enabling many publications that produced new results and/or conclusions from reusing this data, but unfortunately in the current difficult financial climate it did not receive long term funding and currently is not taking new submissions.
Promising moves on the horizon
Despite this setback, things are now more optimistic in this area, with the community looking more receptive to sharing their data, and work underway to overcome many of the roadblocks that have been holding things back (see our commentary on this subject). There are a number of promising moves from brain atlas, (e.g. the Allen Brain Atlas), and connectome projects (e.g. the human connectome project and CMRR) for providing huge resources of public data. In the functional imaging space, the openMRI project also provides a home for raw fMRI data, and has developed nice platform and data organisation standards.
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) was launched in 2003 to speed up drug development by validating imaging and biomarker data for Alzheimer’s disease clinical treatment trials, and set a new standard in neuroscience for data sharing without embargo. Despite data being de-identified, the consortium allows access only to approved members of the “scientific community” after their authorization, and there are stricter rules regarding data reuse and credit than the genomics community are used to. In contrast to these limitations we issue all of our data use a CC0 waiver to maximise its potential re-use, whilst giving all our hosted datasets a citable DataCite DOI to enable attribution and credit to the producers. The broader imaging community has shown that there are mechanisms and a large potential user base for sharing their data, with the Open Microscopy Environment (OME) community producing a suite of open visualization and conversion tools and data standards. OME’s open imaging platform OMERO (developed in part by our editorial board member Jason Swedlow), has a growing user base from labs organizing and presenting their data, and has worked with journals and societies to produce products such as the Journal of Cell Biology image viewer, and the American Society of Cell Biology “The Cell: an Image Library”. In the longer term the future is looking bright, the European Euro-Bioimaging initiative is also planning and aiming to build large-scale public biological and biomedical imaging infrastructure by 2017.
Data Publishing to the rescue?
Complementing these forthcoming subjects specific resources, the rise of a number of unstructured repositories such as Dryad and Figshare gives additional options and makes data sharing even easier. Structuring the data and storing necessary metadata to use it is essential to enable its reuse, and on top of providing the storage infrastructure the time and effort to carry this from data producers out needs to be credited. Data-publishing provides an incentive for data producers to make this effort, crediting early release of data with a citable publication, and allowing publication of the downstream analyses later.
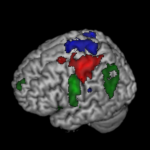 Our recently published data note by Chris Gorgolewski and colleagues is a great example of this, providing test-retest functional MRI data for motor, language and spatial attention functions for 10 de-identified patients. This allows validation of fMRI tasks used in pre-surgical planning for tumor resection, and provides a valuable resource for the development of new methods and algorithms. On top of the GigaScience datanote the authors have already published an analysis using this, demonstrating that this model works. Overcoming some of the previous concerns about deposition of this type of data, all patient details have been deidentified, and the data is structured in the openfMRI manner. These authors are keen proponents of data-sharing, and have talked about the benefits of this type of data-publishing in the past, so it is great to see them use GigaScience as a vehicle for this.
Our recently published data note by Chris Gorgolewski and colleagues is a great example of this, providing test-retest functional MRI data for motor, language and spatial attention functions for 10 de-identified patients. This allows validation of fMRI tasks used in pre-surgical planning for tumor resection, and provides a valuable resource for the development of new methods and algorithms. On top of the GigaScience datanote the authors have already published an analysis using this, demonstrating that this model works. Overcoming some of the previous concerns about deposition of this type of data, all patient details have been deidentified, and the data is structured in the openfMRI manner. These authors are keen proponents of data-sharing, and have talked about the benefits of this type of data-publishing in the past, so it is great to see them use GigaScience as a vehicle for this.
Data publishing is currently very topical, and coming on top of the many other recent schemes we have written about in the past, Nature Publishing Group have recently announced their own foray into the field, with the upcoming launch of Scientific Data. Due to launch in spring 2014, their “data descriptors” are very similar to our data note articles, and also use the interoperable ISA-TAB metadata format that a number of our submitters have utilized. It is flattering that two years after we started publishing data in this way that Nature will be following what we have done, and it encourages and validates our approach, spurring us to take more data. We would data producers to take advantage of the fact that all article and data processing charges are currently covered by BGI until the end of the year, and contact us if you are interested in publishing data notes with us or submit here. For more on our data notes also see here.
Image credits: Charlie Llewellin, Flickr; Gorgolewski et al.
Further Reading
1. Gorgolewski KJ, Storkey A, Bastin ME, Whittle IR, Wardlaw JM, Pernet CR. A test-retest fMRI dataset for motor, language and spatial attention functions. Gigascience. 2013 2:6. http://dx.doi.org/10.1186/2047-217X-2-6.
2. Gorgolewski, KJ; Storkey, A; Bastin,ME; Whittle, IR; Wardlaw, JM; Pernet, CR (2013) A test-retest functional MRI dataset for motor, language and spatial attention functions. GigaScience Database http://dx.doi.org/10.5524/100051
3. Gorgolewski KJ, Storkey AJ, Bastin ME, Whittle I, Pernet C. Neuroimage. Single subject fMRI test-retest reliability metrics and confounding factors. 2013 69:231-43. http://dx.doi.org/10.1016/j.neuroimage.2012.10.085
4. Breeze JL, Poline JB, Kennedy DN. Data sharing and publishing in the field of neuroimaging. Gigascience. 2012 1:9. doi: http://dx.doi.org/10.1186/2047-217X-1-9
Recent comments
Comments are closed.
[…] datasets, and new types of data to our repository. On top of our first neuroimaging data (see the blog posting) and two new rat genomes, we have just added our first two proteomics datasets. Having recently […]
[…] the effects of various genetic defects on the spontaneous activity of the retina. We’ve written previously about the difficulties in sharing neuroscience data, and to date there are not many publicly […]
[…] Neuroinformatics Coordinating Facility (INCF). For example, as our editor Scott Edmunds noted in a blog about neuroscience data notes and our journal GigaScience, one tool promoted by the INCF has been the fMRI data […]
fMRIDC did a great job in the past, but due to difficulties in getting funded it has not been available for over a year. I’m sure INCF acknowledges the important role fMRIDC played in the early years of datasharing, but I doubt it is recommending it right not (since fMRIDC has not been accepting submission for quite a while). Luckily the http://openfmri.org is taking over the baton accepting task based fMRI datasets. I recommend can recommend it for all authors wanting to deposit their task based fMRI data (this is what we did as well https://openfmri.org/dataset/ds000114).