
Our ability to control nature is one of those unique human traits that have allowed us to rapidly and exponentially increase our carrying capacity. By selecting desirable traits on wild organisms extracted from their natural environments, we transformed the human condition from a primarily subsistence lifestyle to one that maximizes our quality of life through high-yielding livestock and crops, and designer pets.
Many of these domesticated traits are quite specialized: certain cow breeds have been selected for higher milk yield, heirloom chickens for outlandish pompadours, and dog breeds for tending to our other domesticates.
However, a handful of traits emerge in common and, together, make up a converged class of phenotypes known as the ‘domestication syndrome’. In particular, behavioral traits such as tameness and docility are shared derived features of domesticated animals and result from generations of adaptation to their new human environment.

Model organisms bred in our laboratories may not be so different
As a human commensal, the fruit fly, Drosophila melanogaster, is thought to have co-migrated with humans, similar to how ancestral canines followed prehistoric humans and scavenged our leftovers.
Over a century ago, these pesky insects were chosen as genetic models to exploit their short generation times and rearing ease, and were immediately put to heavy laboratory use by such early geneticists as Thomas Hunt Morgan and Alfred H. Sturtevant (see right).
Ever since, D. melanogaster has become an indispensible lab model used to address nearly all biological questions.
Are there differences in temperament between wildtype and wild-caught fruit flies? Yes. It is well known among Drosophila geneticists, particularly wine drinkers who regularly interact with natural populations of D. melanogaster, that lab strains are far less active than their wild counterparts.
And catching an escaped Drosophila reared in the lab takes relatively little skill when compared to capturing flies buzzing around empty bottles of Merlot.
Yet, to our surprise, no one had previously reported activity differences between lab-reared and wild-caught flies in the literature.

So, our first step was to quantify these differences using a high-throughput arena and MATLAB-based tracking software (see right).
Our results (available in Supplementary Material) confirm that lab strains are significantly less active and interactive with other flies.
Are these differences due to adaptation to laboratory conditions or are they the result of mutational load?
The conventional belief is that over multiple successive generations of inbreeding, deleterious mutations accumulate independently across laboratory strains under effectively small population conditions. This process would generate a common phenotype similar to the domestication syndrome that many would describe as lethargic.
To address this question, we reassembled five different lab strains that have been isolated in Drosophila laboratories for decades. D. melanogaster also has some of the best population genomic resources available including over 500 fully sequenced lines derived from global populations and a recently updated assembly of its sibling species, D. simulans. Comparing these genomes to the five lab assemblies allowed us to identify derived lab-specific single nucleotide polymorphisms (SNPs) that differentiate lab strains from wild flies.
Comparing these genomes to the five lab assemblies allowed us to identify derived lab-specific single nucleotide polymorphisms (SNPs) that differentiate lab strains from wild flies.
As a whole, the nearly 20K lab-specific SNPs appear to be neutral, and even slightly deleterious. Amino acid changes, codon preferences, and other evolutionary parameters were not much different from randomly placed mutations lending credence to the mutational load hypothesis.
However, when these SNPs were binned according to their frequency across the five laboratory strains, we began to see distinctive patterns in high-frequency SNPs, i.e., found in most or all of the assembled lab strains.
These high-frequency lab SNPs were under greater selective constraints, overrepresented on the X-chromosome (helping to promote recessive traits that easily fix in hemizygous males), and significantly enriched in neurogenetic genes. We also found significantly larger extended haplotypes enriched with these high-frequency derived laboratory SNPs.
Early selection on an appreciable pool of genetic variation
Our best guess is that these high frequency SNPs originated from either an ancestral or ghost population of D. melanogaster. There are far too many lab-specific SNPs that can be accounted by mutations over such a short period of isolation.
In fact, it is believed that a recent global sweep of transposable elements dramatically changed the landscape of D. melanogaster during the last fifty years. Old lab strains, sequestered in laboratories and devoid of these recent mobile genetic elements, provide living fossils to understand how variation has spread in natural populations.
Soon after these lines were extracted from nature from a previously much larger variant pool, slow activity alleles found primarily in X-linked behavioral genes were likely inadvertently favored among the non-escapees. These selected SNPs were then maintained for over a thousand generations in the laboratory while deleterious mutations accumulated via recurrent inbreeding.
How does behavior evolve so quickly?
So, behavior appears to have evolved rapidly after D. melanogaster’s initial shift to the laboratory environment. Dmitri Belyaev’s famous Russian fox experiment also demonstrates that these docile behaviors can evolve across a very small number of generations of domestication.
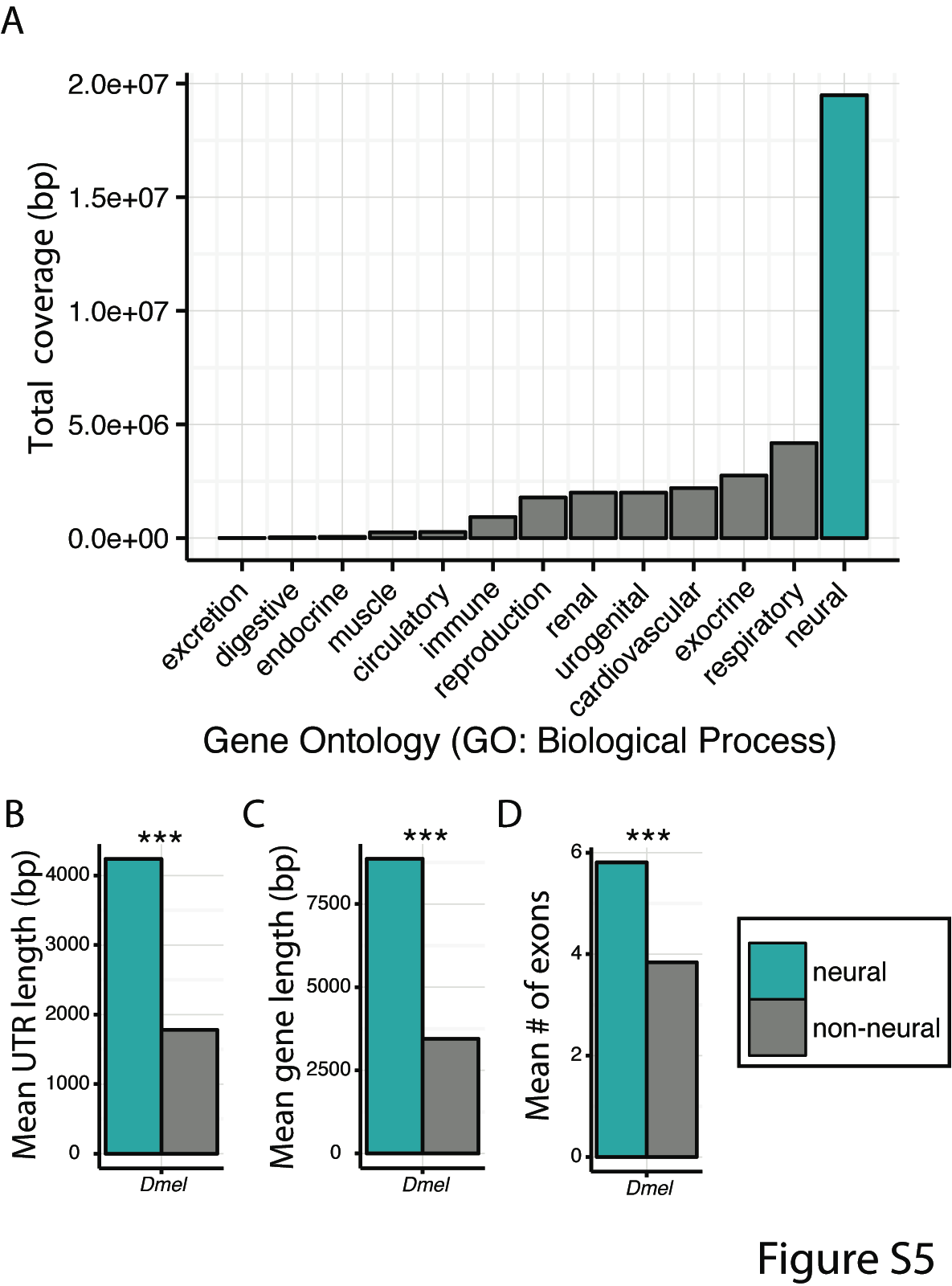
We suggest that genomic architecture may play an important role. In Drosophila, fixed lab-specific SNPs are up-regulated in neural tissue and enriched for neurogenetic and sensory functions, thus, explaining differences in domesticated behavioral phenotypes such as locomotion and activity.
However, when we control for gene length, a number of these enrichments disappear. Is it possible that neurogenetic genes simply provide a larger target for mutations to arise, thus, promoting rapid evolutionary rates of change?
Comparing the gene length of the functional class, ‘neurogenetic’, to other ontological classes from the same hierarchical level, we find large, significant differences (see right) across both regulatory and CDS regions.
This observation suggests a novel genomic mechanism explaining the rapid evolution of common domesticated behaviors in animals.
Larger neurogenetic mutational targets provide a deeper pool of genetic variation for selection to act on, offering an alternative explanation to the behavioral component of the ‘Domestication syndrome’.
In summary…
Our paper suggests that the docile and tame temperament of lab-reared D. melanogaster needn’t solely be explained by the accumulation of deleterious mutations after generations of inbreeding.
We identified adaptive signatures on a subset of lab-specific variants enriched in neurogenetic genes on extended haplotypes that likely have been selected early on in the domestication process and maintained in the bottle, even in the face of accumulating deleterious mutations. Sequencing more of these living fossils will provide greater resolution to understanding adaptation in the laboratory.
We also propose that a large neurogenetic mutational target provides a genomic mechanism for rapid behavioral divergence which may also play a role in the rapid evolution of sexual isolation between incipient species.
Comments