
Coping with complexity: reducing the complexity of network models to make them workable
The size and complexity of many genome-scale metabolic models is growing to feature more and more reactions and metabolites. This means that analyses may be computationally impractical to undertake. In addition, it can be more meaningful to study an organism’s central metabolism with a simpler model, revealing the basic principles of their metabolism.
A new method published in 2015 called NetworkReducer, as its name suggests, reduces large-scale network models to smaller and more tractable networks. These could represent, for instance, the central metabolism or another module of interest. The algorithm protects the most important elements and functions of the network while systematically pruning others.
Suboptimal systems: what’s a cell to do when being optimal is not the ideal?
Flux Balance Analysis (FBA) is one of the staple method of systems biology analyses. It is an approach for working out how metabolites flow through the chain of reactions in a metabolic network to optimize a particular outcome. However organisms don’t always operate optimally, and in the unpredictable environments they find themselves in it may pay them to hedge their bets, keeping their metabolic options open at the cost of not being entirely optimal at present.
A new FBA method called corsoFBA applies this line of thought, predicting metabolism that aims for a less than optimal production of biomass but minimizing the cost in protein required. It was found that often this will differ markedly from the optimal solution. The authors’ simulation results show better agreement with results from some previous experiments in E. coli, than alternative models that assume they were aiming for a goal of optimization.
How bacterial metabolism adapts to spatial location within the colony
The behaviors of individual cells are greatly affected by their surrounding habitat, and the habitat surrounding cells in bacterial colonies will vary greatly depending on their position within the colony: at the edge are pioneers forging out into unknown new territory, whereas life for cells in the middle may be marked by reduced resources and a build-up of waste products.
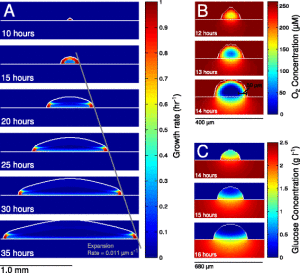
The model described in this paper is of an Eschericia coli colony grown on glucose-minimal agar, using equations to describe the 3D diffusion of nutrients, and metabolic models of the cells show how they would react to the different conditions.
The model reveals a new mechanism of cross-feeding of acetate within the colony, supported by their experimental observations of growing colonies. Outside of their bacterial system, the authors find parallels in this model with growth of tissues and tumors, suggesting there are possibly interesting new dynamics in these too.
Benchmarking challenges for researchers trying to perfect model parameters
Developing and utilizing large-scale mathematical models that predict the dynamic behavior of biological networks is one of the main goals of systems biology. A major component of the effort of reverse engineering these biological models is in estimating the values of the model parameters. They will frequently be unknown and may not even be measurable experimentally, so the aim is to optimize the parameter estimates so as to give the best fit of the model’s predictions to experimental results.
With many researchers working on these problems there needs to be a standardized way to judge their success. BioPreDyn-bench is a set of six benchmark problems for this purpose. The problems posed are in organisms from bacteria to mammalian cells, cover metabolic, signaling and developmental models, and consist of either simulated or measured data. They are provided in standard formats to enable them to be simulated in as many standard tools as possible.
She shapes sea shells: Evolution of shell shapes in ammonites
This article applied a systems biology approach in an area very different to most of the other published articles: the evolution of the form of ammonite shells. These extinct marine molluscs are familiar because their fossilized shells are widely found, and are highly recognizable (and collectable) because of their attractive spiral shape. Although all ammonites had this basic spiral shape, lots of variations in size and shape arose during the ~140 million years of their existence.

One problem evolution faces is that the form of organisms may have several important functions, and it may be impossible to settle on a body shape that optimizes all of these. For example in ammonites one shell form might favor rapid growth but this might trade-off against being economical with shell material or with having a hydrodynamically efficient shape.
This analysis measured thousands of ammonoid species from across hundreds of million years. It found that all the different designs can be boiled down to a small number of features, which can then be plotted to fit into a pyramid-shaped space of possible designs. Each point of this pyramid represents one of five different functions: an organism whose design falls near a point is highly optimized for this function. Following mass extinction events that wiped out many species, over time the same pyramid shape encompassing all species forms evolved again and again, suggesting the basic functions remained constant.
Comments